Figure 1.
Identification of novel AT-binding proteins in ESCs by DNA pull-down mass spectrometry
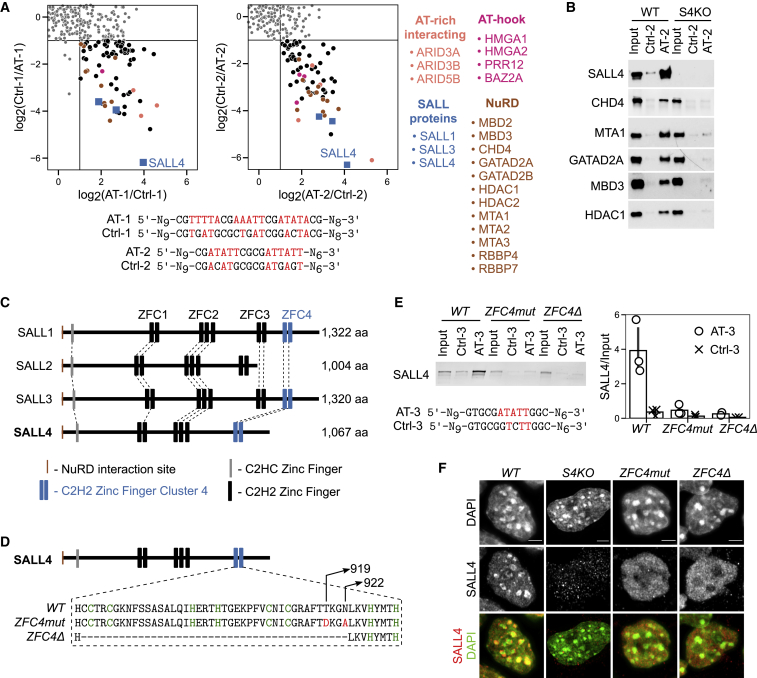
(A) Scatterplots representing SILAC-based DNA affinity purifications, comparing AT-rich DNA probes (AT-1 and AT-2) with control probes having interrupted AT runs (Ctrl-1 and Ctrl-2). The ratio of the quantified proteins in the forward experiment (Heavy-AT/Light-Ctrl) is plotted on the x axis, and the ratio for the same proteins in the reverse experiment (Heavy-Ctrl/Light-AT) is plotted on the y axis. Proteins were considered to be specific interactors when showing at least a 2-fold ratio in both the forward and reverse experiments. These proteins cluster in the bottom right quadrant.
(B) DNA pull-down with AT-rich (AT-2) or control (Ctrl-2) probes followed by western blot analysis for SALL4 and NuRD components using wild-type (WT) or Sall4 knockout (S4KO) ESC protein extracts.
(C) Protein alignment of mouse SALL family members indicating conserved protein domains, including C2H2 zinc-finger clusters (ZFC1–ZFC4).
(D) Diagram showing the mutations or deletion introduced within SALL4 ZFC4 by CRISPR-Cas9.
(E) DNA pull-down with AT-rich (AT-3) or control (Ctrl-3) probe followed by western blot analysis for SALL4 using WT or Sall4 ZFC4mut/Δ ESC protein extracts. SALL4 levels were quantified and normalized to input. Data points indicate independent replicate experiments and error bars standard deviation.
(F) SALL4 immunofluorescence in the indicated ESC lines. DNA was stained with DAPI, showing dense clusters of AT-rich pericentric chromatin. Scale bars, 3 μm.