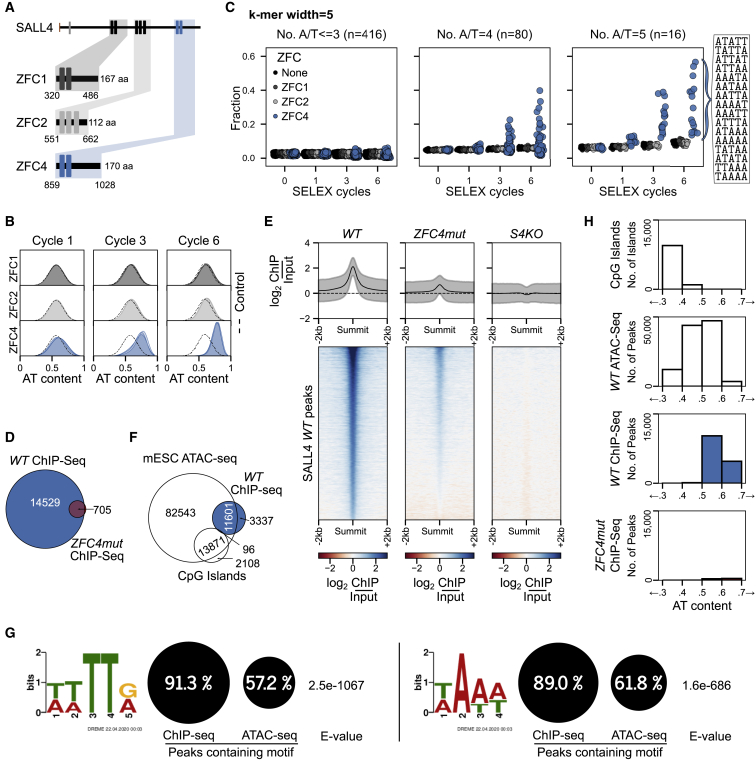
Figure 2.
Characterization of SALL4 C2H2 zinc-finger cluster 4 (ZFC4) DNA binding in vitro and in vivo
(A) SALL4 ZFC1, ZFC2, and ZFC4 protein fragments used for in vitro HT-SELEX experiments. A sample without added protein was used as a negative control.
(B) Base composition of HT-SELEX libraries following 1, 3, and 6 cycles with ZFC1 (dark gray), ZFC2 (light gray) and ZFC4 purified proteins (blue).
(C) Relative enrichment of 5-mer motifs categorized by AT content at cycles 0, 1, 3, and 6 of HT-SELEX with SALL4 ZFC1 (dark gray), ZFC2 (light gray), ZFC4 (blue), and negative control (black) samples. All 16 possible A/T 5-mer motifs are ordered according to their enrichment at cycle 6 of ZFC4 HT-SELEX.
(D) Venn diagram showing the overlap of SALL4 ChIP-seq peaks between WT and ZFC4mut ESCs.
(E) Profile plot and heatmap showing SALL4 ChIP-seq signal at SALL4 WT ChIP-seq peaks in the indicated cell lines.
(F) Venn diagram showing the overlap of SALL4 ChIP-seq peaks detected in WT ESCs with ATAC-seq peaks (accessible chromatin) and CpG islands.
(G) Results from de novo motif analysis at SALL4 WT ChIP-seq peaks (summit ±250 bp) showing the relative frequency of each DNA motif and its associated E-value. ATAC-seq peaks were used as a control for regions of accessible chromatin.
(H) Analysis of the DNA base composition surrounding SALL4 ChIP-seq peaks (summit ±250 bp) in WT (blue) and ZFC4mut (red) ESCs. CpG islands and ATAC-seq peaks coincide with regions of accessible chromatin and are shown for comparison.
See also Figure S2.