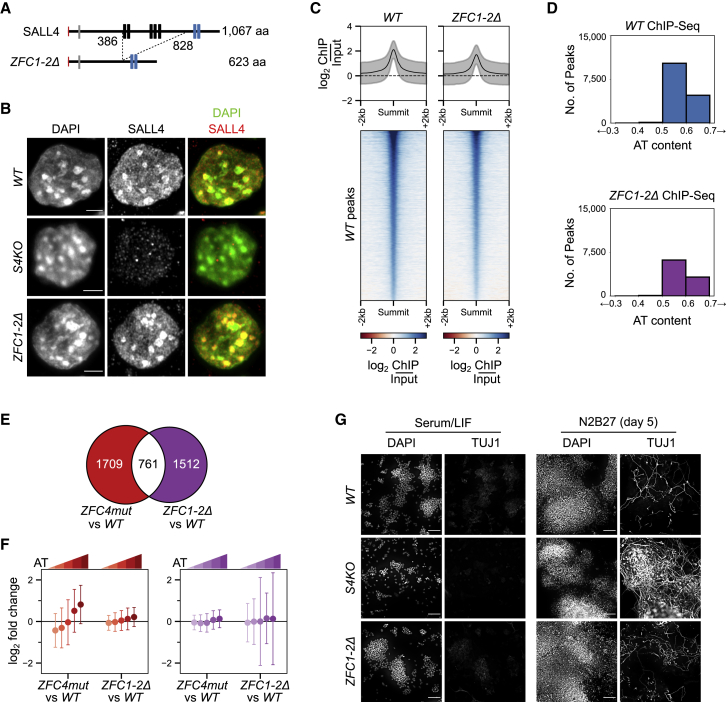
Figure 6.
Effects of SALL4 ZFC1 and ZFC2 deletion in ESCs on chromatin binding, gene expression, and differentiation
(A) Diagram showing the in-frame deletion within the Sall4 coding sequence, generated by CRISPR-Cas9.
(B) SALL4 ZFC1-2Δ localization determined by immunofluorescence in the indicated ESC lines. DNA was stained with DAPI, showing dense clusters of AT-rich pericentric chromatin. Scale bars, 3 μm.
(C) Heatmap and profile plot showing SALL4 ChIP-seq signal at SALL4 WT ChIP-seq peaks in the indicated cell lines.
(D) Analysis of the DNA base composition surrounding SALL4 ChIP-seq peaks (summit ±250 bp) in WT (blue) and ZFC1-2Δ (purple) ESCs.
(E) Venn diagram showing the overlap of differentially expressed genes detected by RNA-seq between ZFC4mut and ZFC1-2Δ ESCs. ZFC4-regulated genes are indicated in red and ZFC1/2-regulated genes in purple.
(F) Correlation between gene mis-regulation (log2 fold change versus WT) and DNA base composition in Sall4 mutant ESCs. ZFC4-regulated (red) and ZFC1/2-regulated (purple) genes were divided into five equal categories depending on their AT content.
(G) TUJ1 immunofluorescence in the indicated ESC lines cultured in serum/LIF medium and following differentiation for 5 days in N2B27 medium. DNA was stained with DAPI. Scale bars, 100 μm.