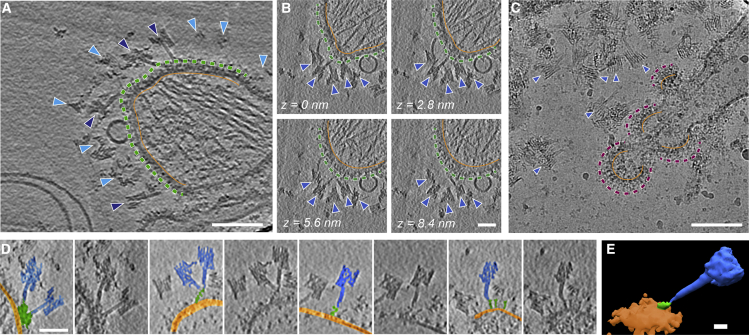
Figure 4.
SPOTs binding to sfGFP-gB vesicles observed by cryoET
Computed tomogram slices in all panels are 2.8 nm thick.
(A) Computed slice from a high-magnification tomogram (1.34 Å/pix) showing SPOTs binding selectively to sfGFP-gB vesicles. The distal edge of sfGFP-gB is indicated by a green dashed line; an orange line is drawn just below the membrane surface. SPOTs are indicated with dark blue arrow heads if their posts are visible in the tomogram slice and light blue if they are situated across multiple slices. Full views of all slices from the tomogram are shown in Figure S4. Average defocus across the tilt series was ∼−3.8 μm. Scale bar, 100 nm.
(B) Sequential computed tomogram slices showing SPOTs bound to sfGFP-gB. Structures are labeled as in (A). Scale bar, 50 nm.
(C) SPOTs are not observed binding to the vesicles in a projection image when gB is tagged with mCherry instead of sfGFP. Magenta dashed lines indicate the distal protein face, and orange lines indicate the inside of the membrane. mCherry belongs to a different family of fluorescent proteins than sfGFP. Scale bar, 100 nm.
(D) Gallery of images of individual SPOTs extracted from tomograms of labeled sfGFP-gB vesicles. Each image is shown twice, with and without annotation on the left and right, respectively. SPOTs are highlighted in blue, sfGFP-gB in green, and the membrane in orange. It is often possible to follow the SPOT posts directly to the recognized protein density on the vesicle surfaces. Scale bar, 50 nm.
(E) A sub-volume average of SPOTs (blue) bound to sfGFP-gB (green) in vesicle membrane (orange) with the orientations determined by the positions of the posts of the SPOTs as initial alignment markers. Scale bar, 10 nm.