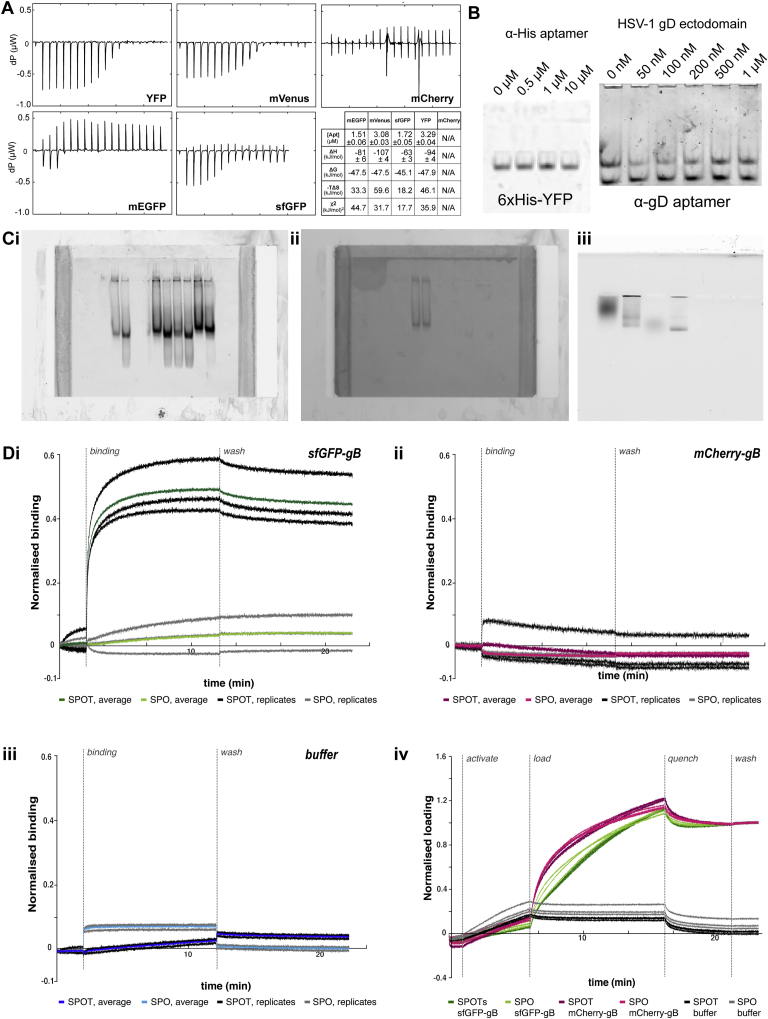
Figure S3.
Biophysical data, related to Figure 3
(A) Raw isothermal titration calorimetry data for fluorescent proteins titrated into aptamer (not linked to origami signpost). The fitted parameters are shown in the table for each binding curve.
(B) We were unable to confirm binding of other aptamers using acrylamide gel shift assays. Left, an aptamer reported against hexahistidine tags (Tsuji et al., 2009); Right, a reported aptamer against glycoprotein D from Herpes simplex virus I (Yadavalli et al., 2017) Ci-iii. Unprocessed gel images with fluorescent detection in the GFP (i), RFP (ii), and YFP (iii) channels for gel-shift assays in Figures 3B and 3C. iii Lane 3 - YFP alone. Because of its low molecular weight (∼26 kDa), we expect YFP to diffuse in the agarose gel, resulting in a band that thus appears faint and with a lower apparent motility. Lane 4 - upon addition of SPOTs to YFP, the molecular weight of the fluorescent structure would increase dramatically when they bind (∼5 MDa) so it diffuses much less, and a sharper and therefore brighter band would be seen. Lane 1 - YFP-Env has a much higher molecular weight than YFP (∼125 kDa monomer, ∼375 kDa trimer). As both forms of YFP-Env are much larger than YFP, the molecules should diffuse less in the agarose and thus appear as a more concentrated band. Additionally, there are 3 YFP on each Env trimer such that we would expect 3x the signal of YFP alone. Lane 2 - upon addition of SPOTs to YFP-Env, the trimer population of YFP-Env could bind 3 SPOTs (∼15 MDa) and the monomer population could bind one SPOT (∼5 MDa). As YFP-Env is a membrane protein produced in mammalian cells, it is glycosylated and detergent-solubilized here, both of which may impact the appearance of bands by electrophoresis. D. Example replicate buffer-subtracted and loading-normalized traces for BLI binding of unfunctionalized sign post origami (‘SPO’) or SPOTs to immobilized sfGFP-gB vesicles (i) or mCherry-gB vesicles (ii) from Figure 3D (bottom panel). The buffer replicates (unloaded tips dipped in analyte nanostructure solutions) and average (used for subtraction) are show in iii, and the loading curves (normalized in the same way as the data) in iv.