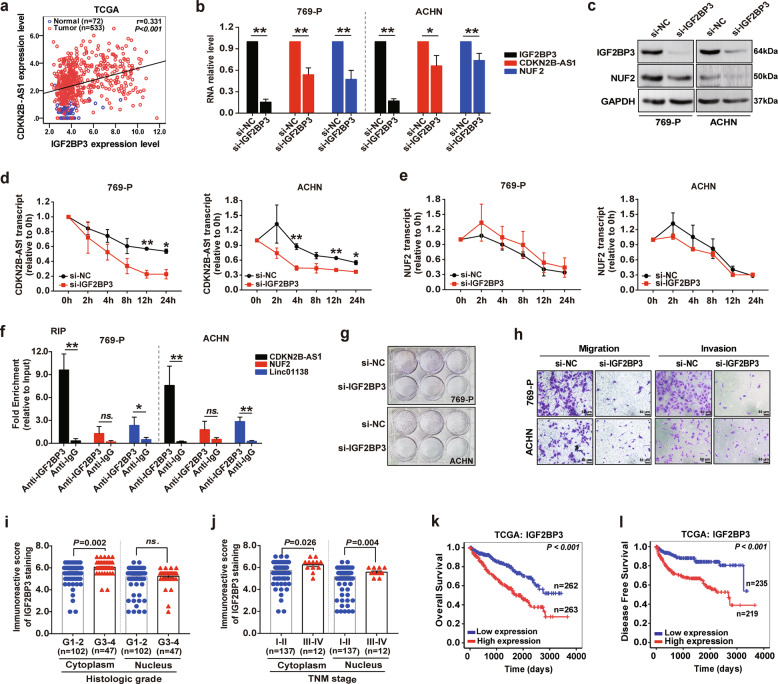
Fig. 5. CDKN2B-AS1 is stabilized by IGF2BP3 in KIRC.
a Pearson correlation analysis of the relationship between CDKN2B-AS1 and IGF2BP3 levels in TCGA-KIRC dataset. Normal tissues are shown as blue circles (n = 72) and tumor tissues as red circles (n = 533). b–e 769-P and ACHN cells were treated with si-IGF2BP3 for 48 h; qRT-PCR analysis of IGF2BP3, CDKN2B-AS1, and NUF2 expression with β-actin as the internal control (b); western blot analysis of IGF2BP3 and NUF2 protein levels with GAPDH as the internal control (c); qRT-PCR analysis of CDKN2B-AS1 (d) and NUF2 mRNA (e) stability, RNA levels were measured after treatment with actinomycin D (5 μg/mL) for 0, 2, 4, 8, 12, and 24 h, respectively, and compared to the level at 0 h. f RIP assay analysis of IGF2BP3 enrichment in CDKN2B-AS1, NUF2, and Linc01138 in 769-P and ACHN cells; RIP enrichment was measured using qRT-PCR normalized by the input RNA. g Colony formation assays were performed to assess cell proliferation in 769-P and ACHN cells treated with si-IGF2BP3. h Transwell assays were used to evaluate migration and invasion in 769-P and ACHN cells transfected with si-IGF2BP3. Scale bar: 50 μm. i, j Immunostaining score of IGF2BP3 in immunohistochemistry of patients with a different histologic grade (i) and TNM stage (j) from the paraffin-embedded KIRC tissue microarray (No. HkidE180Su02). k, l Kaplan–Meier analysis showed the overall survival and disease-free survival in KIRC patients based on the expression of IGF2BP3 in TCGA dataset; the high and low expression groups were divided by the median of IGF2BP3 levels. Error bars represent the SEM from at least three independent experiments, **p < 0.01 vs. si-NC or anti-IgG, *p < 0.05 vs. si-NC or anti-IgG. ns. no significance (p > 0.05). P value calculated by independent sample t test (b–j), or log-rank test (k, l).