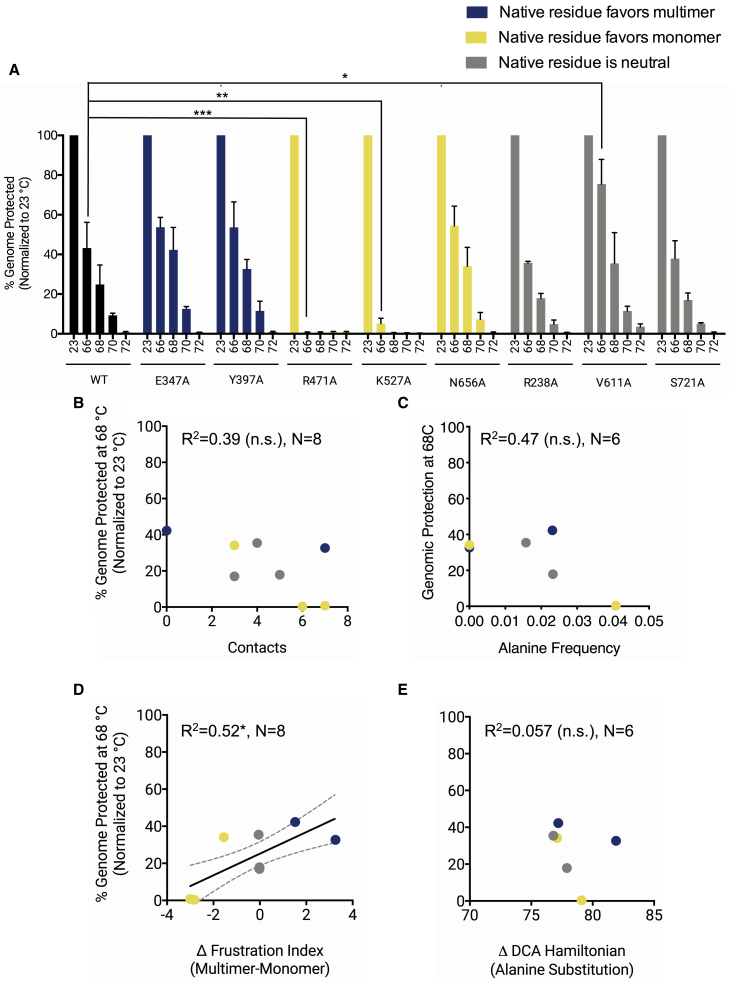
Figure 6.
Thermal stability of alanine mutant capsids. Alanine mutants that formed at sufficient titers were exposed to various temperatures (°C) and then assayed for genomic protection. Temperature-treated samples were incubated with benzonase or sham buffer and genomes quantified using qPCR. (A) Genomic protection after incubation at elevated temperatures is shown. N = 3 independent experiments titered in duplicate. Error bars are mean ± SE. Two-way ANOVA was performed with Dunnett’s post hoc multiple comparison test to compare mutants with WT at each tested temperature. Percentage of genome protected is the fraction of genomes in the benzonase-treated sample as compared with the sham-treated sample. Genomic protection at 68°C is plotted against (B) number of residue-residue contacts in the native capsid structure, (C) alanine frequency in the parvovirus sequence family, (D) Δ-frustration index, and (E) ΔHDCA(Ala). N = 8 mutants for structure-based analyses. N = 6 mutants for sequence-based analyses because these residues do not exist in a majority of the sequence family (these residues are considered as insertions in the hidden Markov model of the family). Regression lines with 95% confidence interval are plotted for statistically significant correlations. R2-values and significance levels are reported. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.