Figure 1.
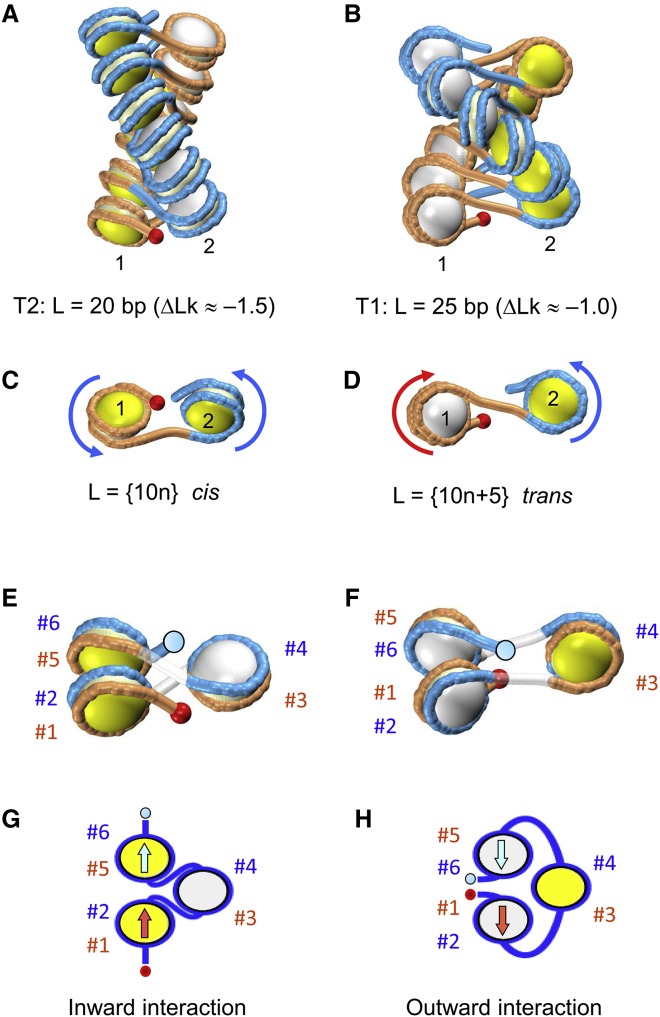
Distinctive “rotational setting” of adjacent nucleosomes in the T2 and T1 topoisomers produces different conformations of the nucleosome arrays. (A and B) Shown are energetically optimal two-start fibers with free DNA linker lengths L = 20 and L = 25 bp, respectively (33). Computations were made for LNCP = 147 bp (59). Note that the red sphere (indicating the “entry” point) is positioned differently in the two fibers; in (A), it is facing the viewer, whereas in (B), it is pointed away from the viewer. In each nucleosome, the entry side is colored in yellow and the exit side in white. (C and D) Shown are the differences in the DNA pathway of successive nucleosomes (here 1 and 2) of the fibers in (A and B). (C) Bottom view, L = 10n is shown. Both nucleosomes face the viewer from the yellow side, and the arrows indicating the DNA trajectory are directed similarly, counterclockwise. The DNA linker contains (approximately) an integral number of helical turns of DNA; thus, the two adjacent nucleosomes are in a cis-like configuration. (D) Front view, L = 10n+5 is shown. Note the different orientations of nucleosomes 1 and 2 that face the viewer from the white and yellow sides, respectively. The arrows run clockwise (nucleosome 1) and counterclockwise (nucleosome 2). Changing the linker DNA length by 5 bp introduces an additional half-turn of the DNA duplex, resulting in a trans-like configuration of the two nucleosomes. (E and F) The entry and exit halves (gyres) of nucleosomes are colored differently to emphasize the distinctive spatial organization of DNA in the T2 and T1 topoisomers. As above, the red spheres indicate the “entry” points and the light blue circles the “exit” points of the trinucleosomes. In (E), gyres #2 and #5 are in contact; we call this the T2 fold. In (F), gyres #1 and #6 are in contact; this is the T1 fold. (G and H) The two types of internucleosome interaction in yeast chromatin observed by Ohno et al. (17) closely correspond to the folding motifs presented in (E and F). Compare the positioning of the “entry” and “exit” points in (E and G) on the one hand and in (F and H) on the other hand. Furthermore, the arrangement of gyres (#1, #2, #5, and #6) is also the same for (E and G) (in the left column) and for (F and H) (in the right column). According to notations used by Ohno et al. (17), the internucleosome contacts shown in (G and H) are denoted “inward” and “outward” interactions, respectively (see their Fig. S1, D and G (17)). Note that the “inward” and “outward” interactions (17) correspond to the IN-IN and OUT-OUT orientations of nucleosomes (according to the notations of Hsieh et al. (14)). To see this figure in color, go online.