Figure 2.
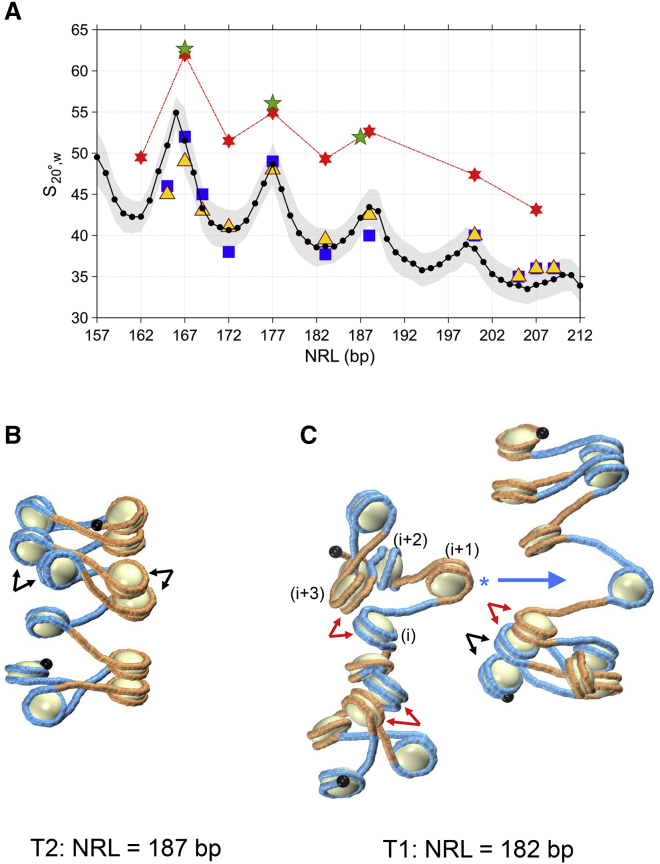
Compactness of nucleosome array folding depends on NRL. (A) Predicted sedimentation coefficient (s20°,w) was calculated for MC ensembles of 12-mer arrays of positioned nucleosomes “601” with LNCP = 146 bp (60). Average MC values are shown by the black solid line, with standard deviations indicated by the gray area (65). The blue squares and yellow triangles represent the experimental values obtained at 1 mM MgCl2 and 150 mM NaCl, respectively (65,72). The red six stars are for the energetically optimal fiber conformations (33) and the green five stars are for the x-ray (51) and Cryo-EM (52) fiber structures, which belong to the T2 family, with L ≈ {10n}. (B and C) Shown are typical MC conformations (65,73) obtained for fibers with NRL = 187 bp (B) and 182 bp (C). Note that the 187 × 12 structure is more compact than the 182 × 12 structures, in agreement with sedimentation experiments (A). Close contacts between nucleosomes are indicated by black (i, i±2) and red arrows (i, i±3). The flipped-out nucleosome (i+1) is marked by an asterisk (C). This nucleosome can interdigitate in the neighboring nucleosome array (large blue arrow). In the fiber with NRL = 187 bp (B), all close contacts are of the (i, i±2) type (shown by black arrows), in agreement with EMANIC data; the nucleosome flipping out occurs much less frequently than for NRL = 182 bp (65). To see this figure in color, go online.