Abstract
Mechanobiology is focused on how the physical forces and mechanical properties of proteins, cells, and tissues contribute to physiology and disease. Although the response of proteins and cells to mechanical stimuli is critical for function, the tools to probe these activities are typically restricted to single-molecule manipulations. Here, we have developed a novel microplate reader assay to encompass mechanical measurements with ensemble biochemical and cellular assays, using a microplate lid modified with magnets. This configuration enables multiple static magnetic tweezers to function simultaneously across the microplate, thereby greatly increasing throughput. We demonstrate the broad applicability and versatility through in vitro and in cellulo approaches. Overall, our methodology allows, for the first time (to our knowledge), ensemble biochemical and cell-based assays to be performed under force in high-throughput format. This approach substantially increases the availability of mechanobiology measurements.
Significance
Here, we have developed a microplate reader assay to encompass mechanical measurements with ensemble biochemical and cellular assays, using a microplate lid modified with magnets. This configuration enables multiple static magnetic tweezers to function simultaneously across the microplate, thereby greatly increasing throughput.
Introduction
Mechanobiology focuses on understanding how physical forces correlate with protein, cell, and tissue dynamics and organization through mechanotransduction (1). It is emerging that mechanotransduction affects almost all cellular processes, from cell-cell and cell-extracellular matrix adhesions to cytoskeletal architecture and gene expression (1,2). In this manner, physical forces provide a mechanism to propagate signals within and between cells (3).
Specialized single-molecule force measurements (such as atomic force microscopy, optical traps, and magnetic tweezers) have been advancing rapidly over the past two decades and have revealed how force is used in biological systems, from individual proteins to complexes and from individual organelles to cells (4, 5, 6, 7, 8). More recently, these techniques have been combined with high-resolution single-molecule fluorescence approaches and cell imaging to visualize dynamic systems under the controlled application of force (9, 10, 11, 12). These measurements have started to probe cellular systems to dissect mechanotransduction pathways.
However, single-molecule manipulation experiments are challenging because of the need for specialized equipment and technical expertise, along with the time to acquire statistically relevant data. Moreover, technical challenges typically limit the experimental design to elegant but minimalized systems, and assays must be optimized for single-molecule conditions. For example, only low concentrations can be used and the reactants must not interfere with the optical detection. Conversely, commonly used ensemble biochemical assays are performed in readily available microplate readers to increase their throughput in determining biochemical constants. However, although offline mechanical measurements have been performed in microplates (13), real-time plate reader assays do not allow mechanical manipulation during measurements and, therefore, are not suitable for performing traditional, established biochemical assays in the presence of force. Therefore, to understand the role of force in biological systems, it is critical to fuse these two approaches.
To this end, we have developed a mechanobiology assay that couples the relative ease of ensemble biochemical assays with magnetic-tweezer-based manipulations to quantitatively study biological processes in the absence and presence of force. This is achieved through the use of magnets incorporated into the lid of a microplate, allowing the assays to be performed in fluorescent microplate readers. Force manipulation of the sample occurs though the tension applied by the magnets to paramagnetic particles attached to the biomolecules of interest. The force applied to the sample can be easily tuned by changing the size of the magnets or adjusting the distance between magnet and sample.
Materials and methods
Materials
Unless stated otherwise, all reagents were purchased from Sigma-Aldrich (Dorset, England). All oligonucleotides were purchased from Integrated DNA Technologies (Leuven, Belgium). Dynabeads were purchased from Thermo Fisher Scientific (Loughborough, UK). MDCC-SSB was prepared as described in (14), BioPcrA was prepared as described in (15) and RepD was prepared as described in (16). BRS-eGFP-Myosin VI Tail-mRFP, NDP52, and GFP-NDP52 was prepared as described in (17).
Different lengths of linear DNA substrates were generated by PCR using the Hot Start Phusion Polymerase (New England Biolabs, Hitchin, UK), as described previously based on the pCERoriD (15). Primers containing 5′ or 3′ modifications for Biotin-TEG or digoxigenin were used to construct the substrates.
BRS-mRFP-Flagelliform(8∗GPGGA)-eGFP, BRS-mRFP- Flagelliform(8∗GPGGA)-HaloTag, BRS-eGFP-Flagelliform(8∗GPGGA)-HaloTag, and BRS-Myosin VI Tail-mRFP synthetic constructs were purchased from GeneArt (Thermo Fisher Scientific, Loughborough, UK). Recombinant constructs were coexpressed with BirA in Eschericha coli BL21 DE3 cells (Invitrogen, Loughborough, UK) in Luria Bertani media supplemented with 50 mM biotin. Proteins were purified by affinity chromatography (HisTrap FF; GE Healthcare, Amersham, UK). The purest fractions were further purified through a Superdex 200 16/600 column (GE Healthcare, Amersham, UK).
The UAS-Gal4 reporter U2OS cells, which stably expressed SNAP-hN1-Gal4 and the H2B-mCherry fluorescence reporter sequence, were kindly gifted by Young-Wook Jun at the University of California, San Francisco.
Three-dimensionally printed magnetic lid
The microplate lid was designed using SketchUp (https://www.sketchup.com) and was produced using Co-Polyester on an Ultimaker 3 Printer. An example CAD file can be downloaded from (https://github.com/cptoseland/MagPlate), and measurements are shown in Fig. S1. The 5-mm cube Neodymium N42 magnets (supermagnete, Gottmadingen, Germany) were attached to the pedestals.
Calculation of force exerted by the magnetic lid
The force exerted upon a sample is dependent upon the size of particle, the gap between magnetic poles, and the distance between magnets and particle. The force is calculated using the equations below, based on the analysis of (18).
For a 2.8-μm particle with a 2-mm gap between magnetic poles,
| (1) |
For a 2.8-μm particle with a 1-mm gap between magnetic poles,
| (2) |
For a 2.8-μm particle with a 0.5-mm gap between magnetic poles,
| (3) |
For a 1-μm particle with a 2-mm gap between magnetic poles,
| (4) |
For a 1-μm particle with a 1-mm gap between magnetic poles,
| (5) |
For a 1-μm particle with a 0.5-mm gap between magnetic poles,
| (6) |
Z refers to the distance between the magnets and particle in millimeters.
Surface modification for in vitro experiments
Glass-bottom microplates (Corning, UK) were used in all assays. Surfaces were treated with 0.2 mg/mL biotin-BSA in 50 mM Tris-HCl (pH7.5) and 50 mM NaCl for 15 min. Surfaces were then washed three times in buffer before adding 20 μg/mL streptavidin and incubating for 15 min. Surfaces were then washed three times with buffer.
Fluorescence spectroscopy
All assays were performed on a BMG Labtech ClarioStar Plate Reader using bottom-optic configuration to facilitate the use of the magnetic lid.
Experimental assays
Förster resonance energy transfer force sensor
Using the prepared surfaces, 100 nM biotin-mRFP-Flagelliform(8∗GPGGA)-eGFP, biotin-mRFP- Flagelliform(8∗GPGGA)-HaloTag, biotin-eGFP-Flagelliform(8∗GPGGA)-HaloTag was added to the surface and allowed to incubate for 15 min at room temperature in 50 mM Tris-HCl (pH 7.5) and 50 mM NaCl. The surface was then washed three times in buffer. 1 mg of Protein-A Dynabeads (2.8 μm) were prepared and loaded with 10 μg of Anti-GFP antibody (ab290; Abcam, Cambridge, UK) or Anti-HaloTag (G9281; Promega, Southampton, UK) according to the manufacturer’s instructions. 10 μg of Dynabead-Antibody fusion was added to the surface and allowed to incubate for 30 min at room temperature. Surfaces were then washed three times in buffer. Fluorescence spectra corresponding to GFP and RFP were then recorded at 25°C in the absence and presence of magnetic lid.
Data was processed to calculate relative Förster resonance energy transfer (FRET).
| (7) |
A is acceptor intensity (RFP, 610 nm) and D is donor intensity (GFP, 510 nm).
Force-induced conformation changes in myosin VI
Using the prepared surfaces, 100 nM biotin-eGFP-Myosin VI Tail-mRFP or biotin-Myosin VI Tail-mRFP was added to the surface and allowed to incubate for 15 min at room temperature in 50 mM Tris-HCl (pH 7.5) and 50 mM NaCl. The surface was then washed three times in buffer. 1 mg of Protein-A Dynabeads (2.8 μm) was prepared and loaded with 10 μg of Anti-RFP antibody (ab62341; Abcam) according to the manufacturer’s instructions. 10 μg Dynabead-Antibody fusion was added to the surface and allowed to incubate for 30 min at room temperature. Surfaces were then washed three times in buffer. Fluorescence spectra corresponding to GFP and RFP were then recorded at 25°C in the absence and presence of magnetic lid or in the presence of 10 μM NDP52. Data were processed to calculate relative FRET using Eq. 7. For titrations, the fluorescence at 605 nm was measured during titration of GFP-NDP52. Background fluorescence arising from direct excitation of RFP was subtracted from the 605 nm intensity. The titration curves were fitted using Eq. 8:
| (8) |
[GFP]t and [RFP]t are the total concentrations for each NDP52 and MVI, respectively. [Complex] is the concentration of the MVI-NDP52 complex.
Surface-immobilized PcrA helicase assays with linear DNA
The streptavidin-coated surface was first treated with 10 nM bioPcrA in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 3 mM MgCl2, and 1 mM DTT for 30 min. One milligram of MyOne Streptavidin C1 Dynabeads (1 μm) was prepared according to the manufacturer’s instructions. 1 μg 1500 bp DNA substrate was incubated for 15 min at room temperature. Beads were washed three times in buffer. A total of 30 μg of Dynabeads were added to each well. A total of 500 nM RepD was then added to the well and incubated for 10 min. The unwinding was initiated with 1 mM ATP, supplemented with 100 nM MDCC-SSB. The reactions were performed at 25 C and MDCC fluorescence (ex. 430 nm and em. 470 nm) was monitored in real-time in the presence and absence of the magnetic lid.
To calibrate the fluorescence signal, three different lengths of DNA 500, 1000, and 1500 bp were used. The helicase assays were performed using the concentrations above but without surface immobilization or magnetic beads. The end point fluorescence signal in solution was then recorded after 5 min. This time should enable complete unwinding of the DNA substrate. The fluorescence signal can then be plotted against DNA basepairs to provide a signal calibration. Figures were made using PlotsOfData (19).
MDCC-SSB interactions with surface-immobilized dsDNA
A total of 1 mg of Protein G Dynabeads (2.8 μm) were prepared and loaded with 10 μg of Anti-Digoxigenin antibody (ab6212; Abcam) according to the manufacturer’s instructions. The Dynabead-Antibody was incubated with 1 μg of 1500 bp DNA substrate for 15 min at room temperature. The streptavidin-coated surface was treated with an equivalent of 2 nM biotinylated 1500 bp DNA containing a 3′ digoxigenin for 15 min at room temperature in 50 mM Tris-HCl (pH7.5) and 150 mM NaCl. The surface was then washed three times with buffer. The concentration 100 nM MDCC-SSB was then added to the DNA and allowed to incubate for 10 min at room temperature. The MDCC fluorescence (ex. 430 nm and em. 470 nm) was then recorded in the presence of the magnetic lid.
Live-cell experiments with the UAS-GAL4 reporter system
The UAS-Gal4 reporter U2OS cells, which stably expressed SNAP-hN1-Gal4 and the H2B-mCherry fluorescence reporter sequence, were cultured in McCoy 5A medium, supplemented with 10% FBS, 50 units/ml of penicillin, and 50 μg/ml of streptomycin and were maintained in a humidified incubator at 37°C and in 5% CO2. Expression of the Notch receptor was induced by incubation with 2 μg/mL doxycycline in cell culture medium for 24 h, before use. Before measurements were taken, the cells were first labeled with 5 μM biotin-conjugated SNAP-tag substrate in tissue culture medium by incubation for 30 min at 37°C and 5% CO2. Cells were washed three times with medium and further incubated for 30 min in fresh medium at 37°C and 5% CO2. Medium was further replaced and supplemented with 30 μg of MyOne Streptavidin C1 Dynabeads (1 μm) per condition. Cells were incubated for 30 min and then recording of mCherry fluorescence was performed in plate reader incubated at 37°C with 5% CO2, over a period of 14 h, in the presence of the magnetic lid. Because of potential cell-to-cell variation, the scan area was expanded to a 2-mm-diameter circle consisting of 30 measurements. The fluorescence signal was subtracted based on starting intensity.
Graphics
Unless stated, data fitting and plotting was performed using plots of data (19) and Grafit Version 5 (Erithacus Software).
Results and discussion
Assay microplate design
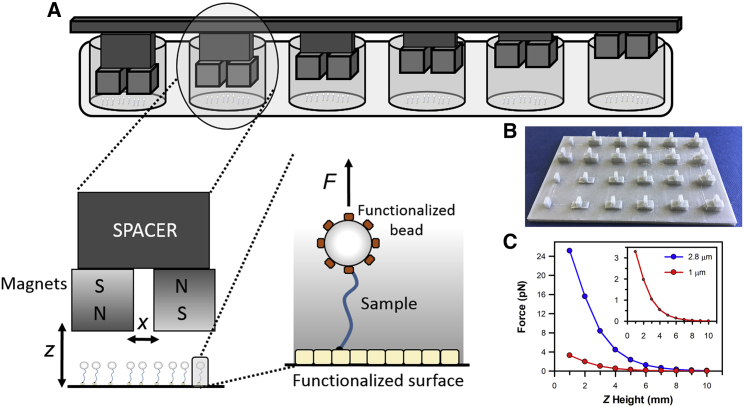
The assay is based around the use of neodymium magnets incorporated into a three-dimensionally (3D) printed microplate lid and biological samples coupled to paramagnetic particles, essentially mimicking multiple magnetic tweezers across the microplate (Fig. 1, A and B). Using a 24-well microplate as an example, a magnet configuration consisting of two 5-mm cube magnets, as used in a magnetic tweezer assay, can be incorporated onto the lid and inserted into each of the microplate wells (Fig. S1). The applied force is dependent upon 1) the distance between the magnets and the sample, which can be varied by the size of spacers within the lid (Fig. 1 A); 2) the distance between the magnets; and 3) the size of the paramagnetic particles used (18). Therefore, by manipulating these properties, we can apply various levels of force across the microplate. Then, we can calculate an apparent force applied to the biological sample on the basis of the known properties’ values (18), as demonstrated in Fig. 1 C. The accuracy of the 3D printing, in terms of the gap between the magnets and the spacer height, is assessed in Fig. S2. A 0.5-mm error in printing the gap between the magnets (labeled x in Fig. 1 A) can yield an average 35% error in force exertion. Likewise, a 0.5-mm error in the spacer height leads to an average 25% error. The 3D printer used here has an accuracy of 0.02 mm, so these defects are unlikely to occur. Further uncertainty can arise because of the variability in the size of commercially available paramagnetic beads. The term-apparent force is used to differentiate the absolute force measurements performed under single-molecule manipulation conditions. This difference occurs because of the inclusion of multiple attachments to a bead within our ensemble data, which would lead to lower exerted forces, versus the selection of only single-bead attachments, which occurs in a microscope. Moreover, because of the close proximity of surface-immobilized molecules, there could be a bias toward multiple attachments unless bead immobilization occurs before adding to the surface. However, assuming the same number of single and multiple attachments would occur in each microplate well, this is a consistent error in our estimation of exerted force.
Figure 1.
For a Figure360 author presentation of this figure, see https://doi.org/10.1016/j.bpj.2020.12.024
Multiplexed magnetic tweezers in a microplate. (A) Schematic of the assay’s format detailing the position and arrangement of neodymium cube magnets within a microplate-based assay. In this format, paramagnetic particles are coupled to the biological molecules of interest, which are in turn attached to the microplate surface. The position of the magnets above the surface (z axis) is defined by spacers incorporated into the 3D printed lid. Illustration is not to scale. (B) The divider that sits on top of the spacers separates the magnets in the x axis by 2 mm. (C) The force exerted upon the biological sample is defined by the size of the paramagnetic particle, the distance of Z above the sample and the spacing between the magnets in X. The force-distance relationship is plotted for the 1- and 2.8-μm particles used in these studies based on the analysis of (18). The inset shows the high distance and low force range.
The assay is configured to be compatible with a fluorescent microplate plate reader. It therefore allows simultaneous multicolor assays over a broad spectrum of wavelengths, as well as the monitoring of intrinsic fluorophores. Here, the measurements are confined to single point at the center of the well (<0.5 mm) where, as with magnetic tweezers, the magnetic field is uniform (Fig. S3 A). The diverse applications of this approach are demonstrated here by monitoring force-induced enzymatic activity, protein conformation changes, and force-dependent signaling pathways. To assess the force dependence of these readouts, we have varied the strength of the applied magnetic field by controlling the distance between the magnets and the sample, as well as the size of the paramagnetic beads. Overall, we have been able to induce biological responses through the modulation of force while simultaneously observing the response through fluorescent microplate-based assays.
Proof of principle: force modulation of protein folding
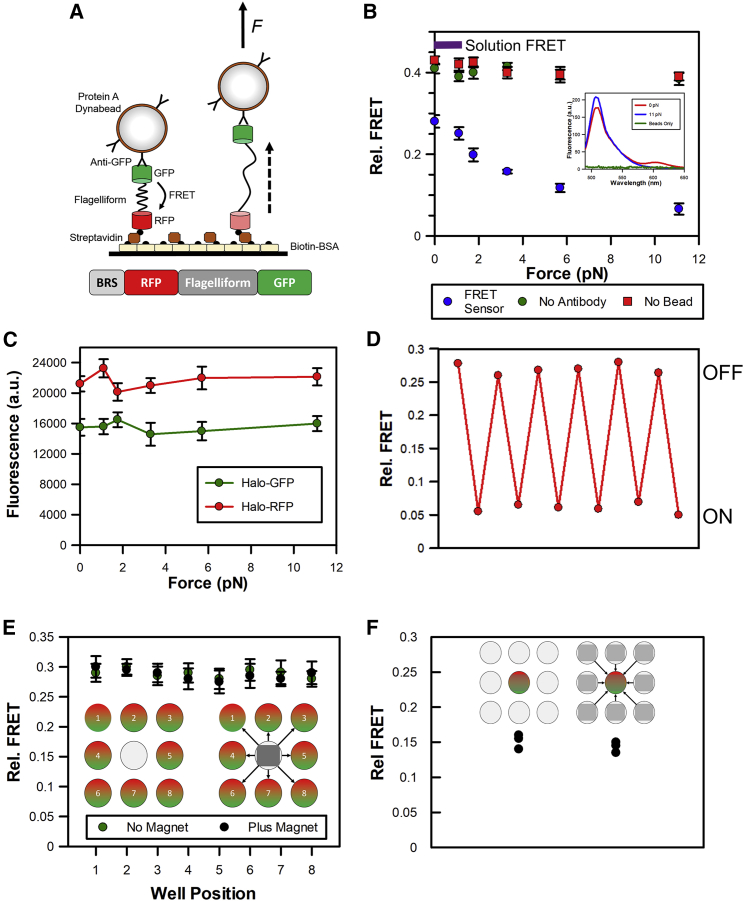
As a proof of principle, we first used a recombinant-FRET-based force reporter (Fig. 2 A). This was constructed by fusing two fluorescent proteins (eGFP donor and mRFP acceptor) to either end of the tension-sensitive peptide repeat from flagelliform, whereby the FRET signal reports upon the force acting across the sensor by unfolding the peptide repeats, as described by Grashoff et al. (20). The sensor also carried an N-terminal biotinylation tag to enable surface immobilization, with its C-terminus coupled to Protein-A Dynabeads through an antibody against GFP. Fluorescence spectra of GFP and RFP were recorded under different experimental formats and the relative FRET signal was calculated from the GFP and RFP emission peaks. The 0 pN of measurement was performed in the absence of magnets and provides a maximal FRET signal in this experimental format (Fig. 2 B). The absence of Dynabeads or antibody resulted in a higher FRET value, suggesting a partial unfolding of the construct resulting from diffusive action on the bead or a change in its fluorescence properties when the antibody binds. Indeed, the FRET signal is similar to that found with the reporter free in solution. Autofluorescence from the Dynabeads had a minor impact on the overall fluorescence signal (Fig. 2 B, inset).
Figure 2.
Proof-of-concept assay format monitoring protein unfolding. (A) Cartoon depicting the assay format monitoring protein unfolding through an eGFP-Flagelliform(8∗GPGGA)-mRFP FRET sensor. The sensor was immobilized on the surface through an N-terminal biotinylation tag and was coupled to Protein-A Dynabeads through an antibody against GFP. (B) Plot of relative FRET signal obtained at different forces. The inset gives an example fluorescence spectra at 0 and 11 pN. A spectrum of the beads alone is also included. Error bars represent SEM from three independent experiments. The purple bar indicates the relative FRET value of the sensor, free in solution. (C) Fluorescence intensity of eGFP-Flagelliform(8∗GPGGA)-HaloTag (510 nm) and mRFP-Flagelliform(8∗GPGGA)-HaloTag (605 nm) obtained at different forces. Error bars represent SEM from three independent experiments. (D) Example data showing alternations between high- and low-FRET states in the presence (ON) and absence (OFF) of 11 pN of force within a single microplate well. The oscillation is consistent with folding and unfolding of the flagelliform repeat. (E) and (F) assess the impact of the magnetic fields upon surrounding microplate wells. (E) Cartoons depict the experimental setup. The FRET sensor was placed in eight wells surrounding an empty central well. The relative FRET signal was then determined for each well. The 11-pN magnet was positioned at the central well and the measurement was repeated. Error bars represent SEM from three independent experiments. (F) Cartoon depicts the experimental setup. The FRET sensor was placed in a central well and exposed to a 6-pN magnet, and the FRET signal was determined. The 11-pN magnets were then placed in the surrounding eight wells and the measurement was repeated. The experiment was performed in triplicate.
In the presence of the magnetic lid (Fig. 2 B), there was an incremental decrease in FRET dependent upon the force applied (Fig. 2 B), which was expected because of the unfolding of the flagelliform peptide and the resulting increase in donor-acceptor distance. The data is in good agreement with the in vitro unfolding experiments performed by Grashoff et al. (20) The previous study showed the peptide fully unfolded, based on loss of FRET, by 8 pN. Those in vitro experiments used cyanine dyes, rather than fluorescent proteins as used here, which may lead to the small differences in unfolding.
The absence of Dynabeads or antibody resulted in no changes in FRET signal in the presence of the magnets, indicating no further unfolding of the peptide. To ensure that the observed FRET signal was not caused by force-induced changes in the GFP or RFP fluorescence intensity due to environmental changes (21), two additional constructs were engineered: biotin recognition sequence (BRS)-mRFP-Flagelliform-Halo and BRS-GFP-Flagelliform-Halo, where anti-Halo antibody was used to couple the constructs to the Dynabeads. Here, the magnet-induced protein unfolding did not cause any intensity changes within the individual fluorescent proteins (Fig. 2 C). Therefore, we are confident that the FRET changes arise solely from protein unfolding. This was further supported by the repeated cycles of folding and unfolding, which were observed by removal and addition of the magnetic lid, respectively (Fig. 2 D).
The use of a microplate offers the advantage of high experimental throughput because a single microplate would accommodate multiple magnetic tweezers conditions compared to a single condition per coverslip in microscopy formats. Our approach is different from strategies for multiplexing magnetic tweezers, which aimed to increase throughput in terms the total number of single-molecule events within a field of view (22). Although these approaches increase the efficiency of performing a single-molecule measurement of a single condition, an experiment still requires multiple coverslips and acquisitions. However, the advantage of our approach could be compromised by the close proximity of the magnets in adjacent wells. To assess if there is cross talk between the magnetic fields of neighboring magnets within different microplate wells, we placed a single magnet pair in the center of the microplate. We then used the FRET reporter to assess the effect of its magnetic field in the surrounding eight wells, which were devoid of magnets. As demonstrated in Fig. 2 E, in the absence of magnets in the central well, the FRET signal in the surrounding eight wells was similar and consistent with the folded conformation of the peptide. Importantly, there is no change in signal when the magnets were placed in the central well, indicating that there is no cross talk between adjacent microplate wells. In addition, we assessed whether the surrounding magnets impact the magnetic field within an individual well. Here, the FRET reporter was placed within a central well and exposed to a single magnet to exert a moderate level of force (∼6 pN) for partial unfolding (Fig. 2 F). The assay was then repeated in the presence of magnets in adjacent wells. Similarly, no difference in the FRET signals was observed, which supports the conclusion that there is no cross talk between the wells.
This methodology is implemented in four types of biological assays to demonstrate the versatility of this tool: 1) force-induced conformation changes in a molecular motor, 2) the unwinding of DNA catalyzed by a helicase, 3) the force-dependent binding of the single-stranded binding protein (SSB) to ssDNA and 4) a force-activated cell-signaling pathway in live cells.
Force-induced conformation changes and ligand binding
As a first application of our approach, we tested its potential in monitoring the force-induced conformation changes within a protein using a similar assay format as above. The actin-based molecular motors, myosins, are regulated by numerous factors, including intramolecular back folding (23). Myosin VI has been shown to exist in an autoinhibited state whereby the association of binding partners, such as NDP52, trigger unfolding and activation of the motor protein (17,24). As a molecular motor, myosin VI is a force-sensing and -generating protein (25,26); therefore, we investigated if forces can also unfold the protein, which may act as a form of regulation.
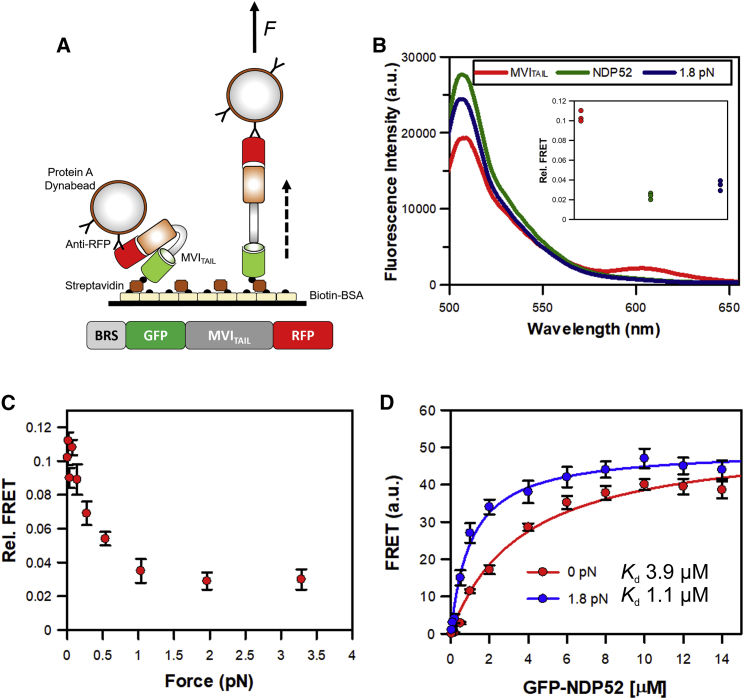
To test this, we used a GFP-Myosin VI Tail-RFP construct, previously used to investigate intramolecular back folding (17). The conformation sensor was immobilized to the microplate surface through N-terminal biotinylation and was coupled to the Dynabeads using an anti-RFP antibody (Fig. 3 A). As shown previously, an FRET population corresponding to the back-folded myosin VI tail was observed and was lost after the addition of 10 μM NDP52, as unfolding occurred (Fig. 3 B; (17)). Interestingly, the presence of 1.8 pN of force also triggered unfolding of the tail domain. When the experiment was repeated for a range of forces, the FRET signal did not change for forces above 1 pN (Fig. 3 C). Although outside the scope of this study, force within this range can be readily exerted upon other cytoskeletal motors through the application of load to the protein. It is therefore likely that force generated by or acted upon the myosin enables unfolding and binding partners can stabilize this conformation.
Figure 3.
Force modulates myosin VI back folding. (A) Cartoon depicting the assay format. The eGFP-MVITAIL-mRFP construct was immobilized on the surface through an N-terminal biotinylation tag and it was coupled to Protein-A Dynabeads through an antibody against RFP. (B) Representative spectra showing GFP and RFP fluorescence in the absence of force upon addition of 10 μM NDP52 or 1.8 pN of force. (C) Plot of relative FRET signal obtained at different forces. Error bars represent SEM from three independent experiments. (D) FRET titration of GFP-NDP52 against BRS-MVITAIL-mRFP under 0 pN (red) and 1.8 pN (blue). The myosin tail was immobilized using biotin and coupled to Protein-A Dynabeads through an antibody against RFP. Fitting the binding curve gave the Kd of 3.9 ± 0.8 and 1.1 ± 0.4 μM for 0 and 1.8 pN, respectively, for the MVI-NDP52 complex.
The overriding benefit of this methodology is the ability to readily measure binding constants modulated by forces. Using the biological system above, we can measure the interaction between NDP52 and myosin VI in the presence and absence of force. Such an experimental approach will allow us to determine if NDP52 can bind to different conformations of myosin VI, which will enable us to dissect the molecular mechanisms governing the myosin regulation.
We modified the assay format to enable FRET to report upon the interaction between NDP52 and the myosin VI tail. Here, we used an RFP tag on the myosin tail and GPF on NDP52. The tail was immobilized through biotin and coupled to Dynabeads by anti-RFP. Titration of GFP-NDP52 in the absence of force yields an equilibrium dissociation constant, Kd 3.9 (±0.8) μM. This is very similar to the 2.1 μM calculated using traditional methods (17,24). The titration was then repeated in the presence of 1.8 pN of force, which is enough to unfold the tail (Fig. 3 B). Binding was once again observed but with a stronger interaction (Kd 1.1 ± 0.4 μM). We can therefore conclude that NDP52 can interact with myosin VI in two conformations but that the unfolded state represents a more stable complex. It is therefore likely that NDP52 will bind to the folded complex and then trigger unfolding of the tail. Alternatively, force exerted by molecular cargo may have already unfolded the myosin and this complex would be stabilized by NDP52 binding.
These experiments represent the strength of this methodology whereby the effect of force on binding constants can be readily assessed.
Force modulation of DNA helicase activity
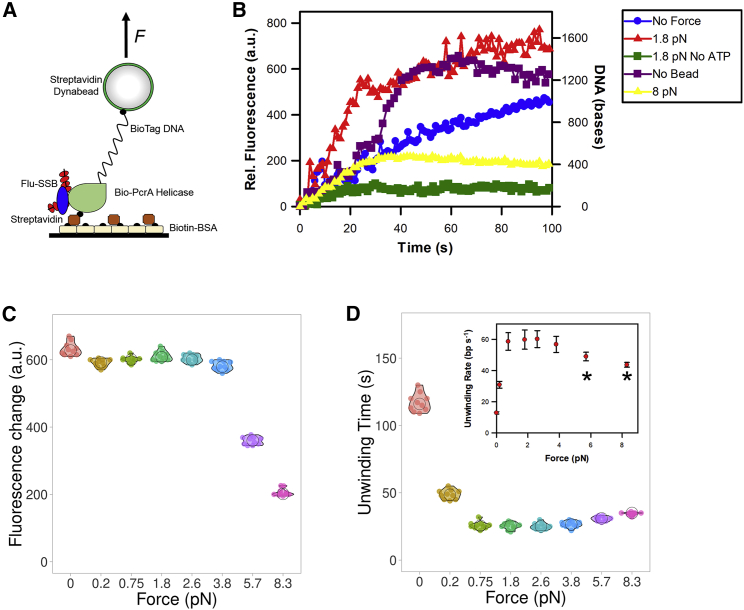
Another application of our approach is to assess the effect of force on the enzymatic activity of a protein. To exemplify this, we used the bacterial DNA helicase PcrA (27). PcrA is a well-characterized ATP-dependent molecular motor, ideally suited for testing the feasibility of our experimental approach and helicases assays can be readily adapted (28). Furthermore, the motor is active when attached to glass surfaces and it can unwind linear DNA fragments (15). As performed previously, we monitored DNA unwinding using a fluorescent ssDNA biosensor (29,30). Biotinylated PcrA helicase was immobilized on optical glass-bottom microplates through biotinylated BSA-streptavidin functionalization (Fig. 4 A). A DNA fragment (1500 bp), with a single biotin tag at the 5′ end, was incubated with RepD to provide a loading site for PcrA helicase. The DNA fragment was then incubated with streptavidin Dynabeads, before blocking the remaining streptavidin binding sites on the bead with biotin-BSA. We used the same functionalization process for both the surface and the beads because they were performed independently.
Figure 4.
PcrA helicase activity on DNA held under tension. (A) Cartoon depicting the helicase-unwinding assay. The helicase was immobilized on the surface, whereas RepD (blue)-loaded DNA was bound to the Dynabead, both through biotin-streptavidin interactions. Fluorescent SSB bound to the ssDNA product to report upon unwinding, which was initiated by the addition of ATP. (B) Representative traces of helicase activity. Helicase-unwinding activity was monitored in real-time by recording the fluorescence change of MDCC-SSB binding to ssDNA. DNA unwinding in the presence of 8 pN of force resulted in a lower fluorescence signal than the other unwinding experiments. Experiments were performed in the absence of Dynabead, magnets (No Force), and ATP. (C) A summary of fluorescence change data for helicase-unwinding experiments performed at the stated force levels is shown. (D) A summary of unwinding time data for helicase-unwinding experiments performed at the stated force levels is shown. The inset portrays the unwinding rates, assuming complete unwinding of the DNA substrate. The black asterisk indicates conditions in which unwinding is incomplete based on (C).
As expected, the immobilized PcrA bound the DNA and then initiated unwinding in the presence of ATP. This was monitored in real-time using the fluorescent ssDNA biosensor, MDCC-SSB (31), whereby the biosensor bound the ssDNA product causing a fluorescence increase, after ATP-dependent DNA unwinding (28). This assay allowed the helicase activity to be followed in the presence and absence of force (Fig. 4 B). In the presence of force, the lid was placed immediately after the addition of ATP.
The fluorescence signal was calibrated against three lengths of DNA (Fig. S4) so that it is possible to measure unwinding in basepairs per second. PcrA was able to unwind the DNA not bound to beads, as observed before (15) with a lag-phase followed by an unwinding phase. PcrA unwound DNA at a rate of 37.5 bp s−1, which compares well to 25–27 bp s−1 using stopped-flow assays and 31 bp s−1 in single-molecule assays (15,16). Unwinding of DNA bound to the Dynabeads reduced unwinding to 12.8 bp s−1. The subsequent addition of 1.8 pN of force led to a fivefold increase in the unwinding rate to 59.8 bp s−1. This is also twofold higher than naked DNA and the trace does not contain the lag period before unwinding began. This suggests unwinding may begin more efficiently. Interestingly, the addition of 8 pN of force led to partial unwinding or a perturbed fluorescence signal. Assuming only partial unwinding occurred, then PcrA functioned at 16 bp s−1.
The addition of Dynabeads impeded DNA unwinding potentially through the addition of greater load on the DNA and/or increased surface interactions. The addition of the magnetic field would elongate the DNA, which would release surface interactions, uplifting the impediment. The overall increase in unwinding rate might reflect a rise in unwinding efficiency because of the elongated, linear DNA geometry. In this manner, the helicase would more readily separate the DNA strands. However, this did not occur when a higher force was applied; instead, unwinding (or at least the measurement of unwinding) was impeded. This may occur because of the displacement of the DNA from the helicase, to a force-induced stalling of the helicase.
To demonstrate the throughput of this assay approach, we performed 10 helicase-unwinding assays at eight different force levels where a measurement at each force level was performed simultaneously. We then calculate the maximal fluorescence change (Fig. 4 C) and the corresponding unwinding time, taken as the time to reach the maximal fluorescent signal (Fig. 4 D). Where the unwinding extent, rather than time, is important, it is possible to run the experiment offline and then scan the entire microplate. This level of throughput cannot be achieved with the corresponding microscopy-based magnetic tweezer assays. We find that each experiment was highly reproducible to within 10%, as seen in Fig. 4, C and D relating to fluorescence change and unwinding time variance. Consistent with the data above, forces above 4 pN led to a decreased fluorescent signal. Moreover, unwinding time decreased as force was applied.
These data highlight the potential of our approach to assess the effect of force on enzymatic activities. Although magnetic tweezer experiments can measure unwinding events in explicit details, this approach can simultaneously record multiple DNA lengths under various conditions (ATP concentrations, salts, other protein factors). Conversely, single-molecule measurements would require multiple coverslips per condition, thereby slowing the throughput and depth of the investigation.
Protein-DNA interactions show a force dependence
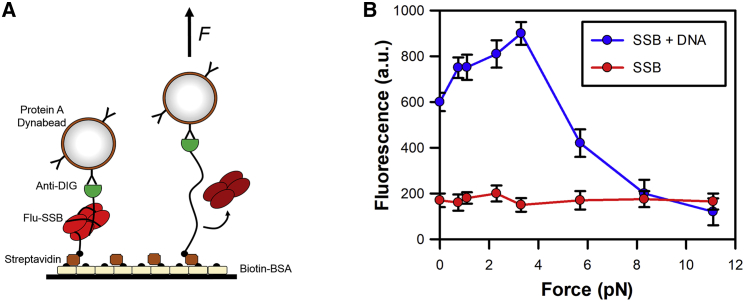
To assess the nature of the SSB-ssDNA interaction, as used above, DNA substrates were generated with a 5′-digoxigenin and 3′-biotin tags to enable coupling of the substrate to the beads and surface, respectively (Fig. 5 A). The PcrA helicase was used to generate the ssDNA before the magnets were applied.
Figure 5.
Force-dependent interactions of SSB protein with ssDNA. (A) Cartoon showing the SSB interaction with ssDNA. ssDNA produced by PcrA unwinding was bound to the surface and paramagnetic bead using biotin and digoxigenin coupling, respectively. Fluorescent SSB was then added to the well and allowed to interact with the DNA. (B) The fluorescence from MDCC-SSB was recorded (at 475 nm) at various forces. Measurements were also recorded in the absence of DNA to provide the background signal generated by free SSB. SSB binding to ssDNA leads to a threefold increase in fluorescence. Error bars represent SEM from three independent experiments.
We have been able to observe that forces up to 4 pN led to an increase in fluorescence signal suggesting there was enhanced SSB binding onto ssDNA (Fig. 5 B). It is possible that the addition of force enables the multiple SSB units bound along the DNA substrate to reorganize, leading to more SSB binding or an alternative-binding mode. However, increasing the force above 4 pN led to a decrease in the fluorescence signal, with background levels being reached at 8 pN. This was consistent with the unwrapping of SSB from the ssDNA in a force-dependent manner, as proposed by Zhou et al. (32). The Zhou et al. study directly measured DNA wrapping around a single SSB unit and not SSB binding itself, therefore we cannot draw parallels to the fluorescence increase observed here.
Force modulation of cell signaling
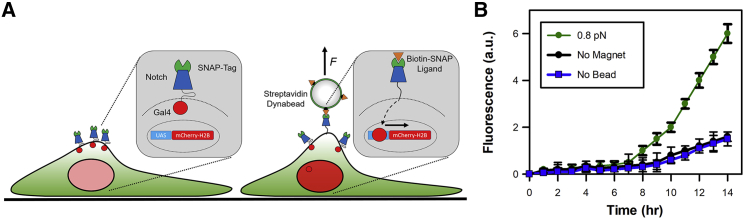
Finally, to demonstrate the applicability of our tool to study mechanotransduction in live cells, we used a U2OS stable cell line expressing an SNAP-tagged human Notch1-Gal4 and a UAS-GAL4 mCherry reporter, which was previously developed for investigating the mechanical response of the Notch receptor (7). In that study, force applied by a magnetic tweezer microscope to the Notch receptor activated downstream signaling, which led to the expression and nuclear localization of the mCherry reporter (Fig. 6 A). We set out to reproduce these experiments in a microplate reader format. U2OS cells were seeded onto a sterile microplate and allowed to grow to 50% confluency. The extracellular region of SNAP-tagged Notch1 receptors were first labeled with biotinylated SNAP ligand and then allowed to attach to streptavidin-coated paramagnetic beads for 1 h, before the magnetic lid was applied. The reporter expression was then monitored by fluorescence for 24 h (Fig. 6 B). The applied force of 0.8 pN led to an increase in reporter expression by 14 h. This expression was fourfold higher than that observed in the absence of magnet or Dynabead, thereby demonstrating mechanical activation of the Notch receptor pathway. We used a lower force than (7) to avoid shearing of the receptor from the cell membrane.
Figure 6.
Force-activation Notch signaling. (A) Cartoon detailing the setup of the live-cell assay to monitor the force-induced activation of the Notch receptor. The modified receptor was coupled to streptavidin-coated paramagnetic beads through an SNAP-biotin ligand. Force across the receptor led to cleavage of the C-terminal domain. In this instance, the C-terminal domain was replaced with Gal4 to act as a transcriptional reporter by driving the expression of mCherry-H2B. (B) The fluorescence emitting from the reporter was monitored in real-time. The fluorescence signal was dependent upon the presence of force and paramagnetic particles. Error bars represent SEM from three independent experiments.
This approach provides a controlled mechanism to activate a receptor in a high-throughput format; therefore, it would be possible to initiate drug screens against such a pathway. No other methodology provides this capability and throughput.
Assay considerations and limitations
Being an ensemble measurement, there are several assumptions made regarding the assay that does not apply to traditional microscopy-based tweezers and single-molecule assays. For example, we cannot determine how many molecules are attached to the beads; therefore, we cannot determine the absolute force exerted on a molecule. However, within the same bead-tether pairing, we assume there is a consistent error between wells on the microplate and therefore we can measure relative force-changes. Premixing beads-tethers before the addition of the surface can reduce the number of multiple attachments because the tethers would not be clustered on a surface.
Moreover, we cannot determine if a molecule is bound by a bead; therefore, we do not determine what fraction of the population of tethers corresponds to generating the signal. However, if bead-tether pairs are formed in solution with a limited concentration of tethers, they can be isolated with magnetic separation. The bound tether-beads can then be added to the surface for tether attachment.
Alternatively, the effect could be examined by varying the amount of beads and surface tethers when setting up the assay to maximize signal change. Importantly, prior knowledge regarding the surface attachment is required to ensure the assay format works with the biological system.
This approach is designed for applying a fixed amount of force. The force cannot be varied within a well and we cannot measure forces exerted by a protein by monitoring bead displacement.
There are also hardware considerations. The microplate reader needs to be able to measure from beneath the microplate with a small scan area. The scan area should be below 1 mm to avoid heterogeneity in the magnetic field. A sensitive detection system is also required to enable to the low concentration (nanomolars) of reactants to be used. Lastly, other magnet types or configurations will need a calibration to be performed to understand the potential force regime on the surface.
Conclusions
In summary, we have developed an easy-to-use, widely applicable tool for ensemble force-based measurements in a microplate format using fluorescent microplate readers. This methodology facilitates mechanobiology measurements in laboratories without the requirement for dedicated single-molecule-imaging facilities. However, we do not have the force accuracy found within single-molecule measurements, and therefore, this approach does not aim to compete with these methodologies. We have exemplified our technology through four different methods representing protein conformation changes, enzymatic activity, protein-ligand interactions, and live-cell reporter assays to demonstrate the versatility of the tool. Overall, this tool can induce biological responses through the modulation of force within this assay format. With this technology, it is now possible to quantitatively study a broad range of biological processes using established ensemble assays in a mechanical context, both in vitro and in cells, or tissues. The approach enables multiple conditions to be tested simultaneously, allowing high-throughput force-induced measurements. Moreover, the microplate design provides high flexibility in the experimental format, ranging from 6-well to 96-well plate, but the latter would require a different magnet format. Therefore, our approach is compatible with biochemical and cell-based assays through to advanced high-throughput drug screens. Furthermore, the microplate design is compatible with all inverted microscopy-based formats, thereby enabling imaging platforms to be combined with mechanical measurements and therefore expanding its range of applicability. This approach substantially adds to the mechanobiological toolbox available to study the impact of force in biological processes.
Author contributions
A.d.S., N.F., and C.P.T. conceived the experiments. A.d.S., N.F., D.S.P., Y.H.-G., and C.P.T. performed experiments and analyzed data. A.d.S., N.F., and C.P.T. wrote the manuscript.
Acknowledgments
We thank the Cancer Research United Kingdom (A26206), the Medical Research Council (MR/M020606/1), and the Royal Society (RG150801) for funding.
Editor: Keir Neuman.
Footnotes
Yukti Hari-Gupta’s present address is MRC LMCB, University College London, London WC1E 6BT, United Kingdom.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2020.12.024.
Supporting material
References
- 1.Jansen K.A., Donato D.M., Koenderink G.H. A guide to mechanobiology: where biology and physics meet. Biochim. Biophys. Acta. 2015;1853:3043–3052. doi: 10.1016/j.bbamcr.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Uhler C., Shivashankar G.V. Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell Biol. 2017;18:717–727. doi: 10.1038/nrm.2017.101. [DOI] [PubMed] [Google Scholar]
- 3.Mohammed D., Versaevel M., Gabriele S. Innovative tools for mechanobiology: unraveling outside-in and inside-out mechanotransduction. Front. Bioeng. Biotechnol. 2019;7:162. doi: 10.3389/fbioe.2019.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dos Santos Á., Cook A.W., Toseland C.P. DNA damage alters nuclear mechanics through chromatin reorganization. Nucleic Acids Res. 2021;49:340–353. doi: 10.1093/nar/gkaa1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elosegui-Artola A., Andreu I., Roca-Cusachs P. Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell. 2017;171:1397–1410.e14. doi: 10.1016/j.cell.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Lherbette M., Dos Santos Á., Schaap I.A.T. Atomic force microscopy micro-rheology reveals large structural inhomogeneities in single cell-nuclei. Sci. Rep. 2017;7:8116. doi: 10.1038/s41598-017-08517-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seo D., Southard K.M., Jun Y.W. A mechanogenetic toolkit for interrogating cell signaling in space and time. Cell. 2016;165:1507–1518. doi: 10.1016/j.cell.2016.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao M., Goult B.T., Yan J. The mechanical response of talin. Nat. Commun. 2016;7:11966. doi: 10.1038/ncomms11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordova J.C., Das D.K., Lang M.J. Combining single-molecule manipulation and single-molecule detection. Curr. Opin. Struct. Biol. 2014;28:142–148. doi: 10.1016/j.sbi.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Madariaga-Marcos J., Hormeño S., Moreno-Herrero F. Force determination in lateral magnetic tweezers combined with TIRF microscopy. Nanoscale. 2018;10:4579–4590. doi: 10.1039/c7nr07344e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newton M.D., Taylor B.J., Rueda D.S. DNA stretching induces Cas9 off-target activity. Nat. Struct. Mol. Biol. 2019;26:185–192. doi: 10.1038/s41594-019-0188-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swoboda M., Grieb M.S., Schlierf M. Measuring two at the same time: combining magnetic tweezers with single-molecule FRET. Exp Suppl. 2014;105:253–276. doi: 10.1007/978-3-0348-0856-9_12. [DOI] [PubMed] [Google Scholar]
- 13.Spencer A., Spruell C., Baker A.B. A high-throughput mechanofluidic screening platform for investigating tumor cell adhesion during metastasis. Lab Chip. 2016;16:142–152. doi: 10.1039/c5lc00994d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook A., Hari-Gupta Y., Toseland C.P. Application of the SSB biosensor to study in vitro transcription. Biochem. Biophys. Res. Commun. 2018;496:820–825. doi: 10.1016/j.bbrc.2018.01.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chisty L.T., Toseland C.P., Webb M.R. Monomeric PcrA helicase processively unwinds plasmid lengths of DNA in the presence of the initiator protein RepD. Nucleic Acids Res. 2013;41:5010–5023. doi: 10.1093/nar/gkt194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toseland C.P., Martinez-Senac M.M., Webb M.R. The ATPase cycle of PcrA helicase and its coupling to translocation on DNA. J. Mol. Biol. 2009;392:1020–1032. doi: 10.1016/j.jmb.2009.07.071. [DOI] [PubMed] [Google Scholar]
- 17.Fili N., Hari-Gupta Y., Toseland C.P. NDP52 activates nuclear myosin VI to enhance RNA polymerase II transcription. Nat. Commun. 2017;8:1871. doi: 10.1038/s41467-017-02050-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Z., Dulin D., Dekker N.H. A force calibration standard for magnetic tweezers. Rev. Sci. Instrum. 2014;85:123114. doi: 10.1063/1.4904148. [DOI] [PubMed] [Google Scholar]
- 19.Postma M., Goedhart J. PlotsOfData-A web app for visualizing data together with their summaries. PLoS Biol. 2019;17:e3000202. doi: 10.1371/journal.pbio.3000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grashoff C., Hoffman B.D., Schwartz M.A. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toseland C.P. Fluorescent labeling and modification of proteins. J. Chem. Biol. 2013;6:85–95. doi: 10.1007/s12154-013-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Vlaminck I., Henighan T., Dekker C. Magnetic forces and DNA mechanics in multiplexed magnetic tweezers. PLoS One. 2012;7:e41432. doi: 10.1371/journal.pone.0041432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fili N., Toseland C.P. Unconventional myosins: how regulation meets function. Int. J. Mol. Sci. 2019;21:67. doi: 10.3390/ijms21010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fili N., Hari-Gupta Y., Toseland C.P. Competition between two high- and low-affinity protein-binding sites in myosin VI controls its cellular function. J. Biol. Chem. 2020;295:337–347. doi: 10.1074/jbc.RA119.010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altman D., Sweeney H.L., Spudich J.A. The mechanism of myosin VI translocation and its load-induced anchoring. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- 26.Hari-Gupta Y., Fili N., Toseland C.P. Nuclear myosin VI regulates the spatial organization of mammalian transcription initiation. bioRxiv. 2020 doi: 10.1101/2020.04.21.053124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 28.Toseland C.P., Webb M.R. Fluorescence tools to measure helicase activity in real time. Methods. 2010;51:259–268. doi: 10.1016/j.ymeth.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Fili N., Mashanov G.I., Molloy J.E. Visualizing helicases unwinding DNA at the single molecule level. Nucleic Acids Res. 2010;38:4448–4457. doi: 10.1093/nar/gkq173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fili N., Toseland C.P., Molloy J.E. A single-molecule approach to visualize the unwinding activity of DNA helicases. Methods Mol. Biol. 2011;778:193–214. doi: 10.1007/978-1-61779-261-8_13. [DOI] [PubMed] [Google Scholar]
- 31.Dillingham M.S., Tibbles K.L., Webb M.R. Fluorescent single-stranded DNA binding protein as a probe for sensitive, real-time assays of helicase activity. Biophys. J. 2008;95:3330–3339. doi: 10.1529/biophysj.108.133512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou R., Kozlov A.G., Ha T. SSB functions as a sliding platform that migrates on DNA via reptation. Cell. 2011;146:222–232. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.