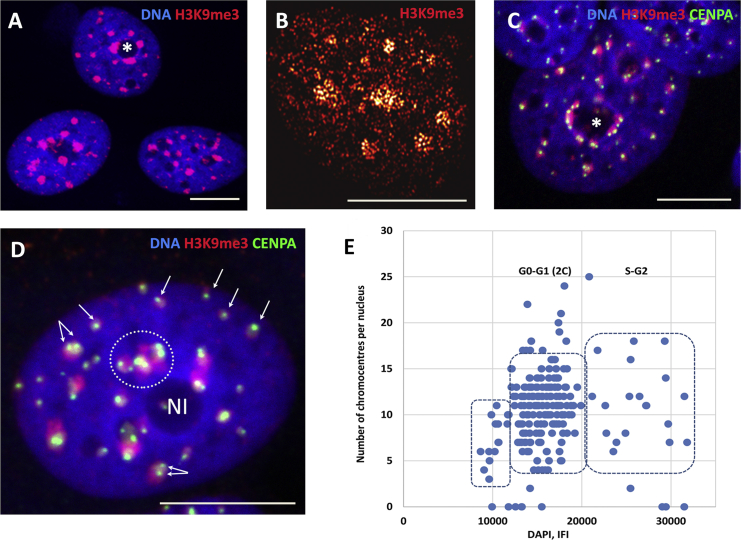
Figure 4.
(A) Epifluorescent microscopy image of a cell nucleus with H3K9me3-positive chromocenters being strongly clustered around nucleoli (asterisk), the nucleus DNA is counterstained with DAPI; (B) H3K9me3-positive chromocenters are represented by high-density regions (bright in the next neighbor image presentation) in single-molecule localization microscopy; (C) confocal section of cell nuclei costained for H3K9me3 and CENPA, identifying the PADs, and the nucleus DNA is counterstained with DAPI; (D) confocal maximum projection image costained for H3K9me3 and CENPA with large PAD clusters (encircled) near nucleolus (Nl) and small PADs (arrowed). Scale bars in panels (A)–(D), 10 μm. (E) H3K9me3 chromocenter numbers versus their size in ST control before adding of HRG; the y axis signifies the number of H3K9me3-positive chromocenters in individual cells, and the x axis signifies the DNA content in these cells as measured by DAPI IFI; the boxed information are as follows: the box to the left shows preapoptotic cells, the box in the center shows the main cluster of 2C cells with highly heterogeneous chromocenter numbers, and the box on the right shows the scarce S-G2 cells.