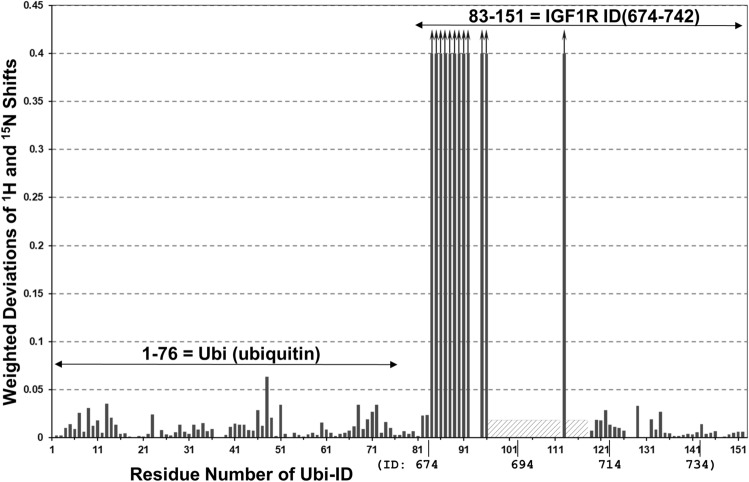
Figure 4.
NMR signal perturbations of the 15N-labelled Ubi-ID protein by VHH-IR5. Weighted deviations are calculated as the square root of the weighted frequency shifts along both the 1H and 15N dimensions of the HSQC spectra of the free Ubi-ID as compared to a 1:1 complex of Ubi-ID with unlabelled VHH-IR5 (Fig. S3a) and plotted according to residue-specific assignments achieved for Ubi-ID (Fig. S4). The ubiquitin moiety from 1 to 76 exhibits relatively small differences between the free protein and its complex with VHH-IR5. Some residues of the IGF1R ID 694-742 moiety show pronounced perturbations, especially for T675-E687 (or residues 84-96 of Ubi-ID). Bars with an arrow indicate those residues whose HSQC signals disappeared in the complex of 15N-labelled Ubi-ID with VHH-IR5. Hatched boxes indicate that no HSQC signals were found for this region of Ubi-ID (R689-R709), except for R704. HSQC signals of the ID segment re-emerge from residue 119 to 151 (R710-E742), with essentially no responses to VHH-IR5 binding.