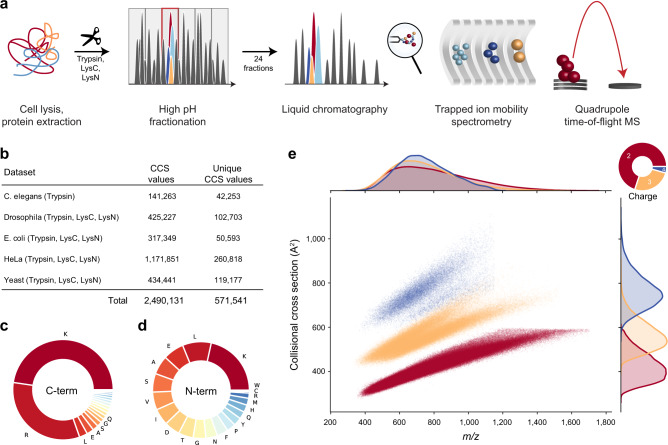
Fig. 1. Large-scale peptide collisional cross section (CCS) measurement with TIMS and PASEF.
a Workflow from extraction of whole-cell proteomes through digestion, fractionation, and chromatographic separation of each fraction. The TIMS-quadrupole TOF mass spectrometer was operated in PASEF mode. b Overview of the CCS dataset in this study by organism. c Frequency of peptide C-terminal amino acids. d Frequency of peptide N-terminal amino acids. e Distribution of 559,979 unique data points, including modified sequence and charge state, in the CCS vs. m/z space color-coded by charge state. Density distributions for m/z and CCS are projected on the top and right axes, respectively. Source data are provided as a Source Data file.