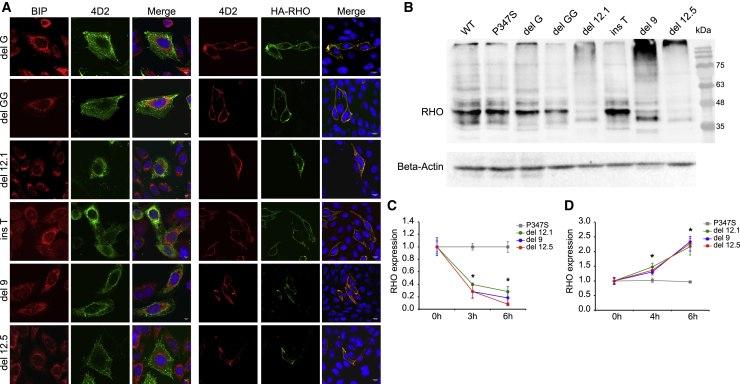
Figure 2.
Biochemical characterization of the most frequent RHO variants generated after editing
(A) Immunofluorescence analysis of CHO cells transfected with plasmids coding for RHO variants. Permeabilized cells (left) were stained with anti-BIP and anti-4D2 antibodies, and the scale bar represents 5 μm. Cells not permeabilized (right) were stained with anti-4D2 and anti-HA antibodies, and the scale bar represents 10 μm.
(B) Immunoblot analysis of WT, p.Pro347Ser, and most frequent RHO variants generated after editing expressed in CHO cells transfected with the respective coding plasmids. Anti-HA antibody was used to detect rhodopsin (RHO). The immunoblotting was normalized with an anti-beta-actin antibody.
(C) Densitometric analysis of immunoblots performed on CHO cells transfected with plasmids coding for p.Pro347Ser, del9, del12.1, and del12.5 RHO variants and treated with 10 μg/mL cycloheximide (CHX). The experiment was performed in triplicate and is presented as mean ± SEM. ∗p value < 0.05.
(D) Densitometric analysis of immunoblots performed on CHO cells transfected with plasmids coding for p.Pro347Ser, del9, del12.1, and del12.5 RHO variants and treated with 50 μM MG-132 proteasome inhibitor. The experiment was performed in triplicate and is presented as mean ± SEM. ∗p value < 0.05.