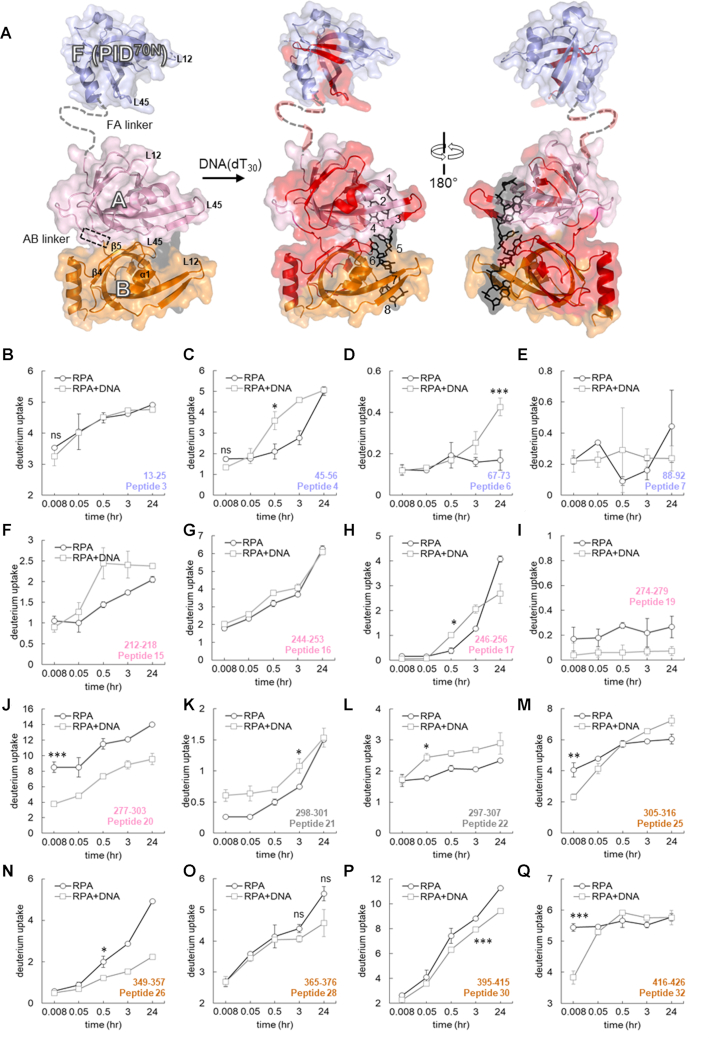
Figure 3.
Time-resolved HDX-MS analysis of DNA-induced conformational changes in the FAB half of RPA. (A). Peptides identified in the MS analysis are mapped onto the crystal structure of PID70N, DBD-A and DBD-B and colored in red. The F-AB linker and A-B linkers are denoted by dotted lines. DNA is depicted as black sticks. (B–Q) Deuterium exchange profiles of individual peptides from the FAB half of RPA. Error bars reflect standard deviations, calculated as described in ‘Material and Methods’ section. */**/*** are P-values calculated using a two-tailed unpaired t-test between the respective time points for the RPA and RPA+DNA samples. Each star denotes t-test values that differ by an order of magnitude. P-values are denoted in Supplementary Table S2. No significance indicated as ‘ns’.