Figure 6.
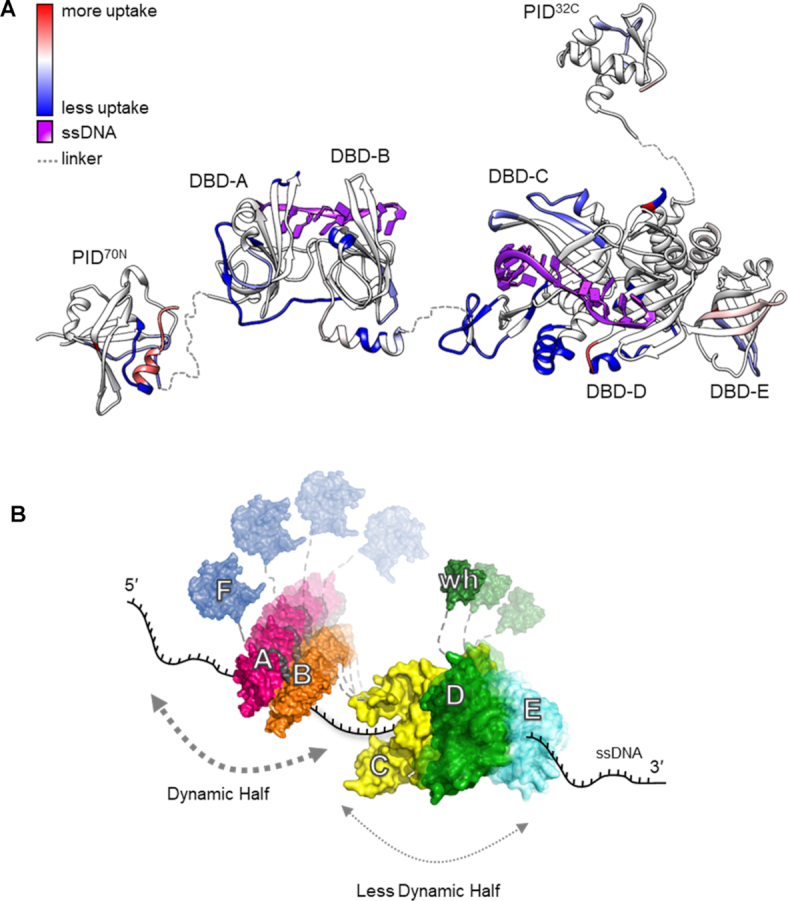
RPA binds to ssDNA using dynamic and less dynamic halves. (A) Global HDX changes in human RPA upon binding to DNA are mapped onto a composite structure. Blue and red denote the difference in scale of deuterium uptake for the unbound (RPA) versus DNA-bound (RPA+DNA) forms of RPA. DNA binding primarily drives protection of regions from deuterium uptake as they become occluded from solvent. (B) An updated dynamics-based model for RPA+DNA interactions is depicted. RPA is proposed to function as two halves (FAB and CDE/Tri-C). PID70N (F domain), DBD-A and DBD-B form the dynamic half and have short residence on time. DBD-C, RPA32 (DBD-D) and RPA14 (DBD-E) form the less dynamic half and have longer residence time on DNA owing to the stability of DNA binding arising from the larger DBD-C domain. The refined model allows RPA to be stably bound to DNA using all DBDs while providing access to both ends of the DNA due to the dynamics of the individual DBDs. PID70N and PID32C primarily serve to bind and recruit RPA-interacting proteins onto DNA.
