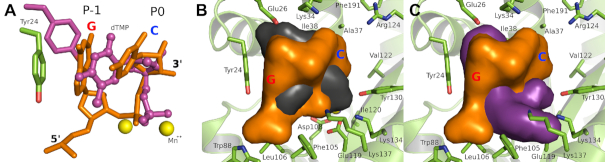
Figure 4.
Implications for PAN endonuclease inhibitor development. (A) Overlap of the bound P-1/P0 RNA dinucleotide with the previously determined bound mononucleotide (pdb file 5CCY). Note that the mononucleotide actually spans the P-1 and P0 sites. The dinucleotide is colored orange and the mononucleotide is colored magenta and shown in ball-and-sticks. The Mn2+ ions at the active site are shown as yellow spheres. Note that Tyr24 in the mononucleotide complex (magenta sticks) moves to maintain the stacking interaction with the base. (B) Spatial comparison of baloxavir and the RNA dinucleotide bound at the P-1 and P0 sites. (C) Spatial comparison of L-742,001 and the RNA dinucleotide bound at the P-1 and P0 sites. In (B) and (C), the RNA dinucleotide is shown as an orange van der Waal surface, the PAN cartoon and key residues are colored lime green, and baloxavir and L-742,001 are shown as black and magenta van der Waal surfaces, respectively. In all panels, the RNA dinucleotide and the PAN endonuclease are derived from the wild type RNA-bound model shown in Figure 3B with GC occupying the P-1 and P0 pockets.