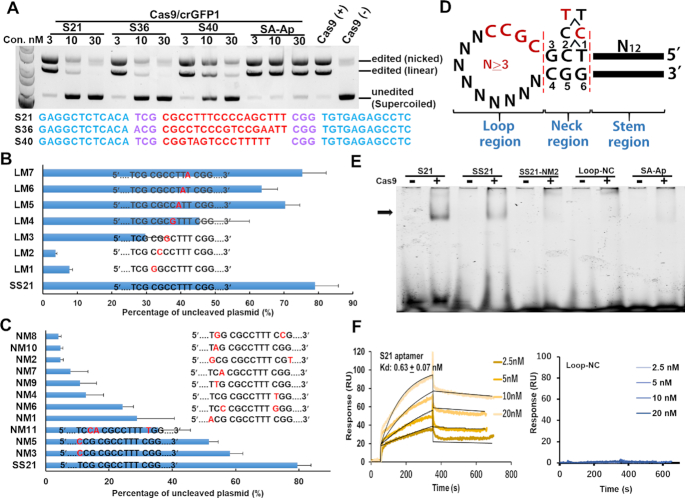
Figure 1.
Analysis of the inhibitory activities of aptamer candidates for Cas9 in in vitro-cleavage assays. (A) Three candidate aptamers (S21, S36, and S40) showed high inhibitory activities for Cas9/crGFP1 at low concentrations. The pCX-EGFP plasmid was cleaved with Cas9/crGFP1 at the GFP1 site in the presence of the aptamer candidate. The mobilities of open circular (nicked), linear and covalently closed circular (supercoiled) plasmids are indicated on the right. SA-Ap: streptavidin aptamer. (B) Mutational analysis of the inhibitory aptamer in the loop region. Thirty nanomolar aptamers were used in the assays. SS21 (a shortened inhibitory aptamer of S21) was used as the parental aptamer. Mutations in the ‘CGCC’ sequence [LM1–LM4, especially at the first or second position (LM1 or LM2)] seriously impaired the capability of the SS21 aptamer. Electrophoresis data for these graphs are presented in Supplementary Figure S2. (C) Mutation analysis of the inhibitory aptamer in the neck region. Aptamers were used at a concentration of 30 nM. (D) Summary of mutation experiments. ‘CGCC’ in the loop region and the sequences in the neck region were invariable regions of S21. The inhibitory activities of mutant aptamers in the neck region were ranked as follows: 5′-T1C2–3>5′-C1C2–3>5′-T1T2–3. (E) Gel mobility shift assay showing the interaction of inhibitory aptamers (S21 and SS21) with Cas9. Loop-NC: negative control where the neck and loop regions of S21 were replaced by tetra-loop, ‘GTAA’. SA-Ap: a streptavidin aptamer. (F) Surface plasmon resonance analysis of the S21–Cas9 interaction. To minimize the mass transport-limited binding effect, a low level of biotinylated inhibitory aptamer or the negative control aptamer was immobilized on the streptavidin chip, and measurements were performed with a high flow rate. To eliminate nonspecific DNA binding to target proteins, the Kd was measured at a high salt condition (250 mM NaCl). Data are presented as means ± SD from three independent experiments. Error bars represent S.D. No response was observed with the Loop-NC negative control.