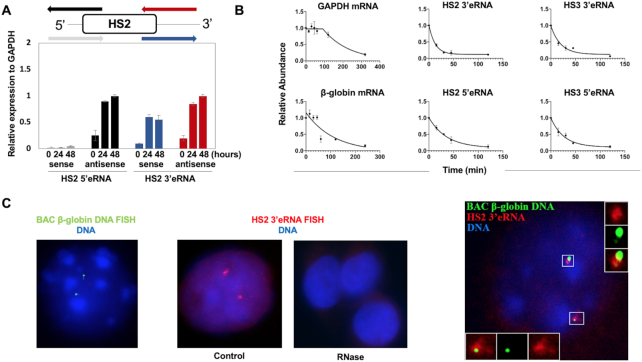
Figure 2.
Formation, stability and localization of LCR associated eRNA. (A) Transcription of 5′ and 3′ HS2 eRNAs during differentiation of MEL cells. Diagram on top showing primers used to assess HS2 5′ and 3′ antisense and sense transcription. MEL cells were subjected to differentiation for 24 and 48 h with 2% (final v/v) DMSO. RNA was isolated, subjected to reverse transcription using strand-specific primers, and analyzed by qPCR. (B) Measurements of GAPDH, β-maj-globin RNA as well as HS2 and HS5 5′ and 3′ eRNA half-life. RNA was isolated from differentiated MEL cells incubated with actinomycin D for up to 8 h and subjected to RT-qPCR using primers specific for GAPDH, β-maj-globin, and HS2 or HS3 5′ and 3′ eRNAs. Error bars reflect SEM from three independent experiments. (C) Differentiated MEL cells were subjected to β-globin gene locus DNA-FISH (left, representative nucleus) or HS2 3′eRNA-FISH (middle-left, representative nucleus; middle-right: RNAse treatment to show specificity of RNA signal, three representative nuclei) using fluorescently labeled probes specific for the murine β-globin gene locus (green) or HS2 3′eRNA (red; DNA: blue). Right: combination of HS2 3′eRNA and β-globin gene locus DNA FISH. HS2 3′eRNA (red) is associated with the β-globin gene locus (green; DNA: blue; four out of four nuclei showed association between the globin gene locus on HS2 3′eRNA).