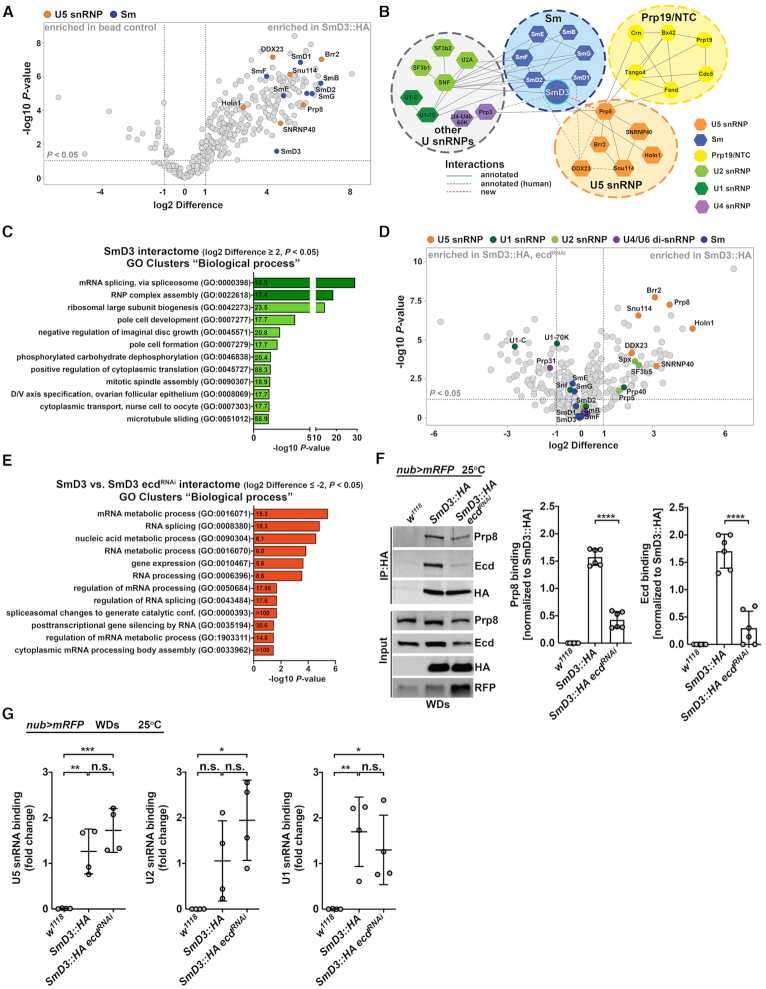
Figure 5.
Ecd deficiency hinders U5 snRNP biogenesis. (A) The volcano plot depicts SmD3-specific interactome identified by LC–MS/MS analysis based on a pull-down of SmD3::HA protein from nub>mRFP, SmD3::HA wing imaginal disc cell lysate. 252 proteins enriched in SmD3::HA IP samples relative to control (nub>mRFP) (|log2 Difference| ≥ 1 and FDR < 0.05) are located within the outlined area. Specific interaction partners including Sm proteins (blue), U5 snRNP proteins (orange) are highlighted. (B) Visualization of a simplified SmD3 protein interactome using Cytoscape highlights its central position within the Sm ring and the spliceosome. Annotated interactions are shown with full or dashed gray lines, new interactions with green dashed lines. (C) The bar graph shows functional GO clusters enriched (fold enrichment ≥ 15, FDR < 0.05, depicted within the bars) among the SmD3::HA interacting proteins (|log2 difference| ≥ 2 and FDR < 0.05). GO terms linked to RNP assembly and RNA splicing are highlighted in dark green. (D) The volcano plot visualizes results of a comparative LC–MS/MS analysis showing differences in protein binding to SmD3::HA precipitated from control (nub>mRFP, SmD3::HA) wing discs lysates and those where ecd was knocked down (nub>mRFP, SmD3::HA, ecdRNAi). RNAi-mediated silencing of ecd transcript reduces interaction of SmD3 with numerous core components of the U5 snRNP (orange) but not with other Sm proteins comprising the Sm ring (blue). The interactions of SmD3 with U1 (dark green), U2 (light green) and U4/U6 snRNP-specific proteins were less affected compared to the components of U5 snRNP. (E) The bar graph shows functional GO clusters enriched (fold enrichment ≥ 5, FDR adjusted < 0.05, depicted within the bars) among the SmD3::HA interacting proteins that are significantly affected (|log2 difference| ≥ 2 and FDR < 0.05) by ecd RNAi knockdown. (F) Representative western blot and quantifications of independent IP experiments corroborate reduced binding of endogenous Prp8 to SmD3::HA in lysates prepared from ecd depleted (nub>mRFP, SmD3::HA, ecdRNAi) wing imaginal discs compared to control nub>mRFP, SmD3::HA samples. Proteins were pulled down with help of the anti-HA antibody-coupled to magnetic beads. Imaginal discs expressing RFP only (nub>mRFP) served to control for unspecific binding. Data represent means ± SD, n = 6. Unpaired two-tailed Student's t-test was used to determine significance, ****P < 0.0001. (G) RIP assays from wing imaginal disc samples using the anti-HA antibody-coated magnetic beads reveal that transgenic SmD3::HA protein precipitates endogenous U5, U2 and U1 snRNAs in the presence (nub>mRFP, SmD3::HA) as well as absence of ecd (nub>mRFP, SmD3::HA, ecdRNAi). Imaginal discs expressing RFP only (nub>mRFP) served to control for unspecific binding. U snRNA levels were determined by RT-qPCR. Data were normalized to the respective input U snRNA levels. Data represent means ± SD, n = 4. Ordinary one-way ANOVA with Tukey's multiple comparisons test was used to determine significance, *P < 0.05, **P < 0.01, ***P < 0.001, n.s. = non-significant. See also Supplementary Figure S5 and Supplementary Dataset S2.