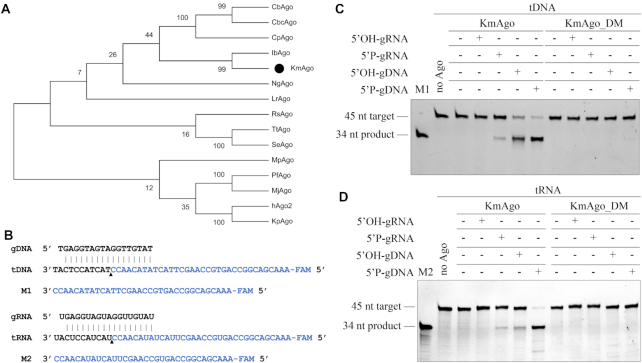
Figure 1.
KmAgo exhibits DNA-guided and RNA-guided RNA cleavage activity at 37°C. (A) Maximum likelihood phylogenetic tree analysis of KmAgo based on amino acid sequences. The numbers at the nodes indicate the bootstrap values for maximum likelihood analysis of 1000 resampled data sets. (B) Sequence of the synthetic let7 miRNA-based guide and target sequences that were used for the in vitro cleavage assays. Black triangle indicates the cleavage site. (C) Cleavage activity assay with FAM-labeled ssDNA target. (D) Cleavage activity assay with FAM-labeled RNA target. KmAgo, guides and targets were mixed in a 4:2:1 molar ratio (800 nM KmAgo preloaded with 400 nM guide, plus 200 nM target) and incubated for 1 h at 37°C. Catalytic mutant KmAgo_DM was used as a control. Lanes M1 and M2 contain chemically synthesized 5′-end, FAM-labeled, 34-nt ssDNA and RNA corresponding to the cleavage products of target DNA and target RNA, respectively.