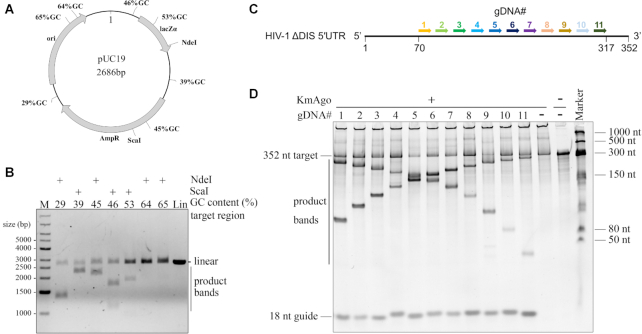
Figure 7.
Double stranded plasmid DNA and highly structured RNA cleavage by KmAgo. (A) Schematic overview of the pUC19 target plasmid. Black polylines indicate target sites while percentages indicate the GC-content of the 80 bp segments in which these target sites are located. (B) Pre-assembled KmAgo-gDNA complexes targeting various pUC19 segments were incubated with pUC19. Cleavage products were incubated with NdeI or ScaI and were further analysed by agarose gel electrophoresis. M, molecular weight marker; Lin, linearized plasmid. (C) Schematic overview of the HIV-1 ΔDIS 5′UTR. Arrows with different colors indicate the target region and the corresponding gDNAs are numbered from 1 to 11 with the corresponding colours. (D) Substrates and products generated by the assay described in (C) were resolved by denaturing PAGE (8%) revealing cleavability of the highly structured RNA by KmAgo–gDNA complex.