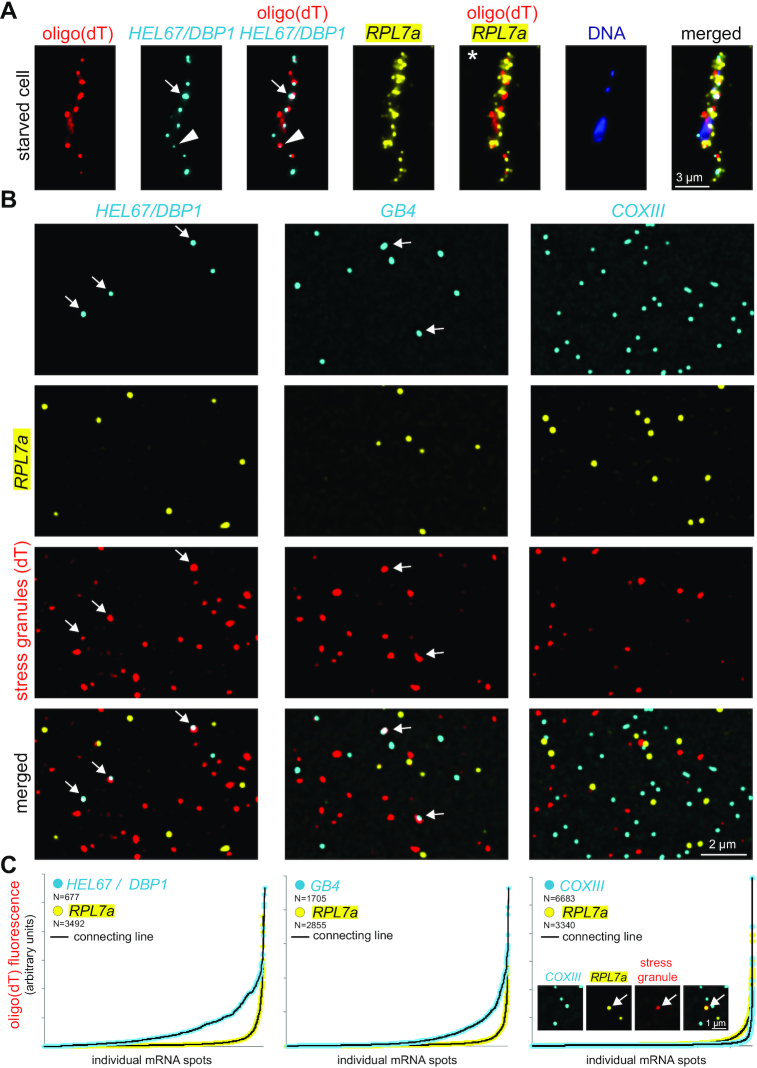
Figure 4.
Detection of mRNAs in stress granules. (A) Starved trypanosomes were chemically fixed and probed for stress granules using an oligo(dT) probe (red) and probes for two specific mRNAs (HEL67/DBP1 shown in cyan and RPL7a shown in yellow). For HEL67/DBP1, a huge variance in the intensity of the different mRNA dots is observed. Often, fluorescence within a granule is low (triangle). Strong, non-spherical signals indicate accumulation of more than one mRNA molecule in a granule (arrow). RPL7a was shown to be underrepresented from RNA granules (6), but its abundance prevents to distinguish whether it is fully excluded from granules or whether a small percentage is granule-localized (asterisk). (B) Samples of LR White embedded starved trypanosomes were probed for stress granules using an oligo(dT) probe (red). In addition, samples were incubated with Affymetrix probe sets for RPL7a (shown in yellow) and either HEL67/DBP1, GB4 or COXIII (shown in cyan). Representative images are shown (projection of a Z-stack of 5 slices with 140 nm distance). (C) Individual mRNA dots were identified (find maxima function of FIJI) and the red fluorescence (oligo(dT) fluorescence = stress granule localization) was measured in each dot. All mRNA dots are shown sorted according to their oligo(dT) fluorescence. The inlet (right graph) shows a rare example of a colocalization between RPL7a and stress granules.