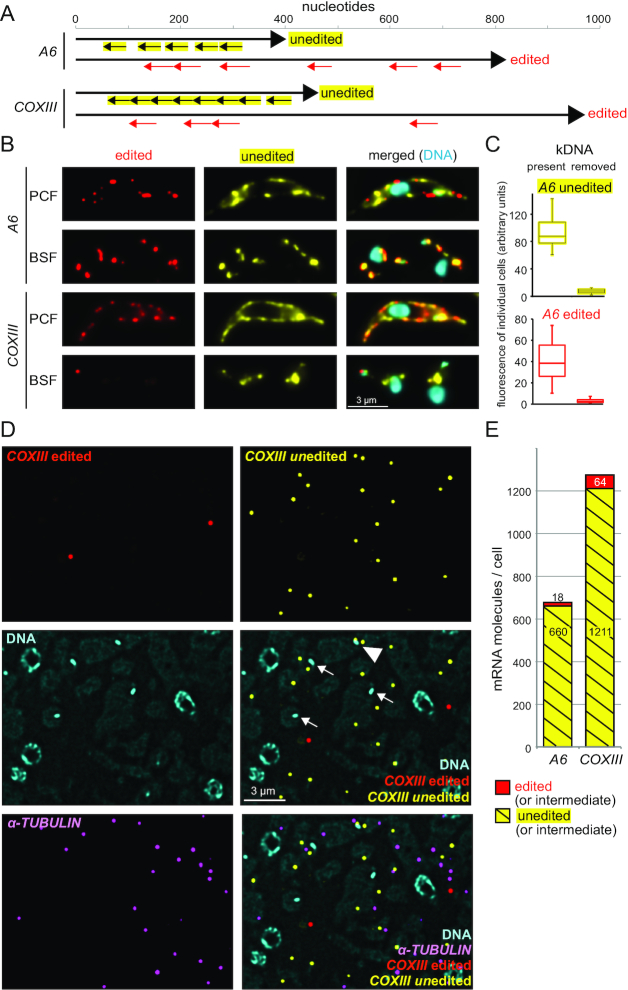
Figure 7.
Detection and localization of edited and unedited mitochondrial RNAs. (A) Schematics of the A6 and COXIII edited and unedited mRNAs showing the positions of the probe pair binding sites. Edited mRNAs have regions of low sequence complexity due to the large number of uridine insertions, and large sections of the mRNA had to be omitted from probing or even blocked with special blocking oligos (further details on the probes are in Supplementary Table S1 and Supplementary Figure S5). (B) Chemically fixed BSF and PCF cells were probed as indicated. One representative cell is shown (projection of a deconvolved Z-stack with 50 slices a 140 nm). For further images, see Supplementary Figure S6, S7A and S8A. (C) To obtain additional evidence for the specificity of the probe sets, we used chemically fixed BSF cells that lacked kDNA and therefore cannot produce mitochondrial transcripts. Such akinetoplastic trypanosomes can be produced by treatment with 10 nM ethidium bromide for three days (42,43) and, if cells carry the compensating mutation L262P in their ATPase gamma subunit, they can survive this treatment (44,45). Control cells and akinetoplastic trypanosomes were probed for edited and unedited A6, and as a control, for the cytoplasmic mRNA FUTSCH. We observed a mild reduction in the number of FUTSCH dots per cell (from 1.7 ± 1 to 0.9 ± 0.9; N > 220), possibly caused by the minor effects 10 nM ethidium bromide have on nuclear DNA. To compensate for this effect, we only included cells with exactly two FUTSCH dots each in the further analysis. The fluorescence of edited and unedited A6 signal was measured from ten individual cells with exactly two FUTSCH dots and the (background-corrected) data are presented as boxplot (waist is median; box is interquartile range (IQR); whiskers are ±1.5 IQR). Akinetoplastic cells have, on average, 13 and 14 times less fluorescence of unedited and edited A6, respectively, than control cells. Microscopy images are shown in Supplementary Figure S7. The corresponding dataset for COXIII is shown in Supplementary Figure S8. (D) LR White embedded PCF cells were probed for edited and unedited COXIII and for α-TUBULIN (as a marker for cytoplasmic mRNA). One representative image is shown as deconvolved Z-stack projection (5 slices a 140 nm). Most mRNA molecules are not close to the kinetoplasts (which are marked by arrows). One rare exception of an unedited COXIII molecule close to a kinetoplast is marked with a triangle. Images of samples probed for A6 are shown in Supplementary Figure S9. (E) The numbers of mRNA molecules per cell for both edited and unedited A6 and COXIII mRNAs were calculated as described above (Figure 3). The total number of nuclei used for quantification was 5841 and 2909 for A6 and COXIII, respectively.