Figure 3.
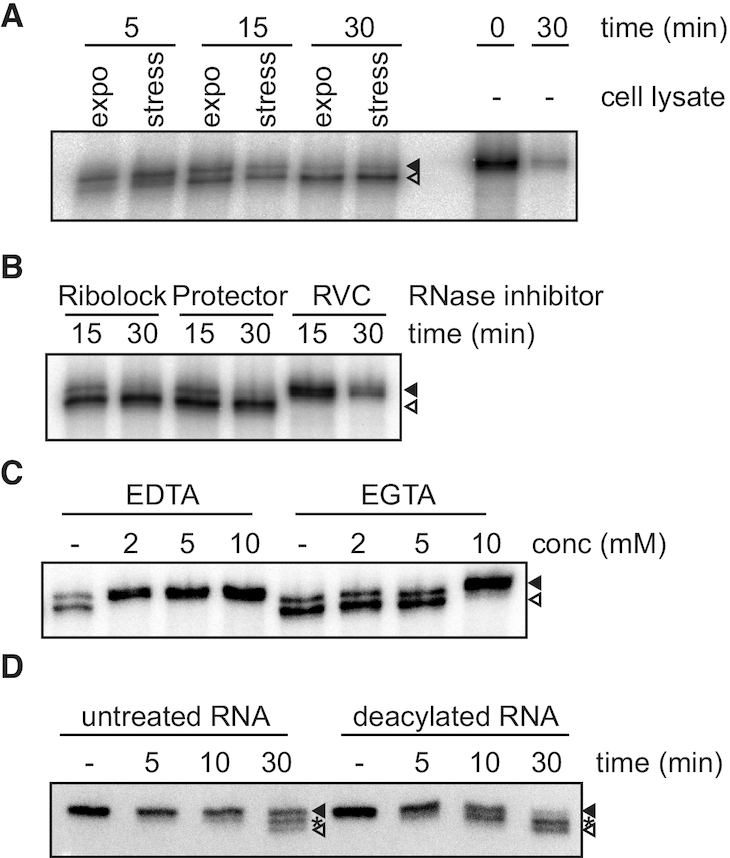
The nuclease trimming the tRNAs is constitutively expressed in cells and can be inhibited by ribonucleoside-vanadyl complex and Mg2+ chelators. (A) The tRNA trimming activity was tested in vitro by incubating total cell lysates from cells grown exponentially (expo) or nutritionally stressed (stress) with 5′ 32P-labeled tRNAVal. After the indicated times total RNA was precipitated, separated on 8% polyacrylamide/urea gels, dried and the radioactivity detected by exposure to phosphoimager screens. Incubation of the substrate under the same conditions in the absence of cell lysate (−) serves as control. (B, C) Cell lysates were treated with specific RNase inhibitors (Ribolock, Protector or RVC; B) or different concentrations of divalent cations chelators (C) and the cellular tRNA trimming activity tested as before. (D) The effect of aminoacylation on tRNA trimming was tested using RNA extracted from cells expressing a tagged tRNAGlu. After incubation of total cell lysates with total RNA either aminoacylated (untreated) or subjected to alkaline deacylation RNA was extracted and tRNA shortening investigated by northern blot against tRNAVal. Full length tRNAVal and fully 3′ trimmed tRNAVal are indicated with filled and open arrowheads, respectively. Asterisks indicate intermediate cleaved tRNAVal.
