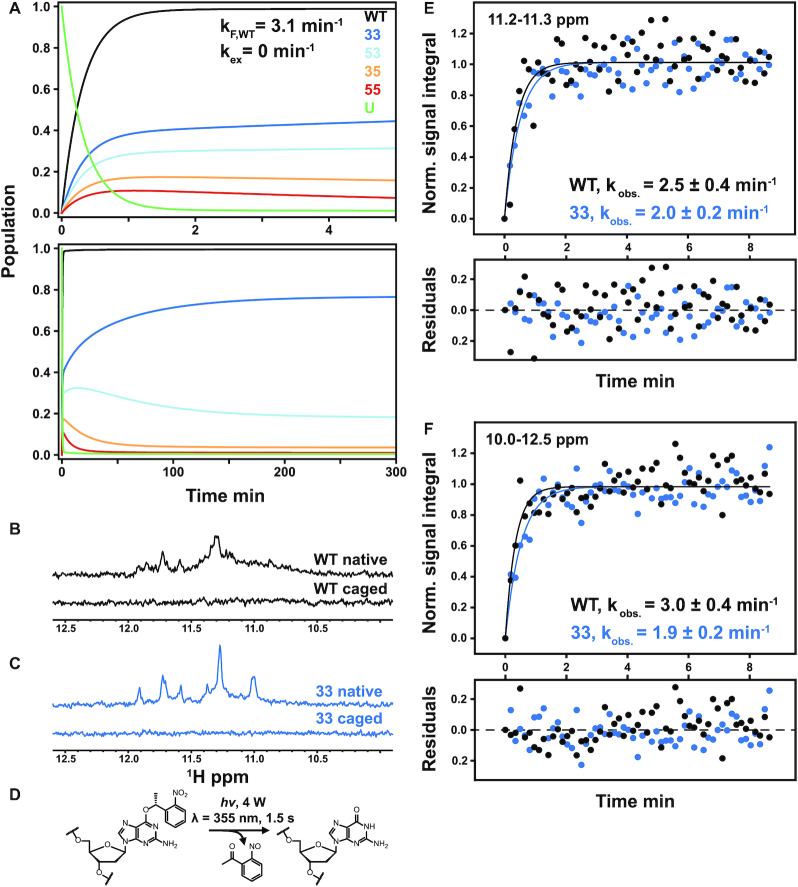
Figure 5.
Theoretical prediction of folding acceleration in the c-myc Pu22 WT ensemble by TH and experimental validation by isothermal NMR folding experiments. (A) Short- and long-timescale isothermal population distributions predicted by TH are shown in the top and bottom plots, respectively. U (green) denotes the unfolded state, and the WT trace is the sum of the profiles for the four GR isomers (colors matching Figures 2 and 3). Isothermal simulations were performed according to Scheme 1C using the optimized parameters from Table 1 without direct GR isomer interconversion (37 °C and kex = 0 min−1, see Supplementary Methods for details). (B and C) Imino proton region of the 1D 1H spectra of 3x-NPE-caged and native (after irradiation) c-myc Pu22 oligonucleotides: WT (B) and 33 fully trapped mutant (C) at 37 °C. For the 3x-caged oligonucleotides, the absence of imino proton signals indicates an unfolded state with no stable Hoogsteen base-pair interactions. (D) Photolysis reaction for O6-(R)-NPE-caged dG with laser light at 355 nm. Folding is initiated after the caging group is released from the dG residue. (E and F) Normalized kinetic NMR data for the light-induced folding of photocaged c-myc Pu22 WT and 33 fully trapped G4s using imino proton signal integration limits of 11.2–11.3 and 10–12.5 ppm, respectively. Data (colored points) were fit to a monoexponential function 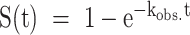 (colored lines) with fit residuals shown underneath each panel. Extracted rate constants are indicated.
(colored lines) with fit residuals shown underneath each panel. Extracted rate constants are indicated.