Figure 1.
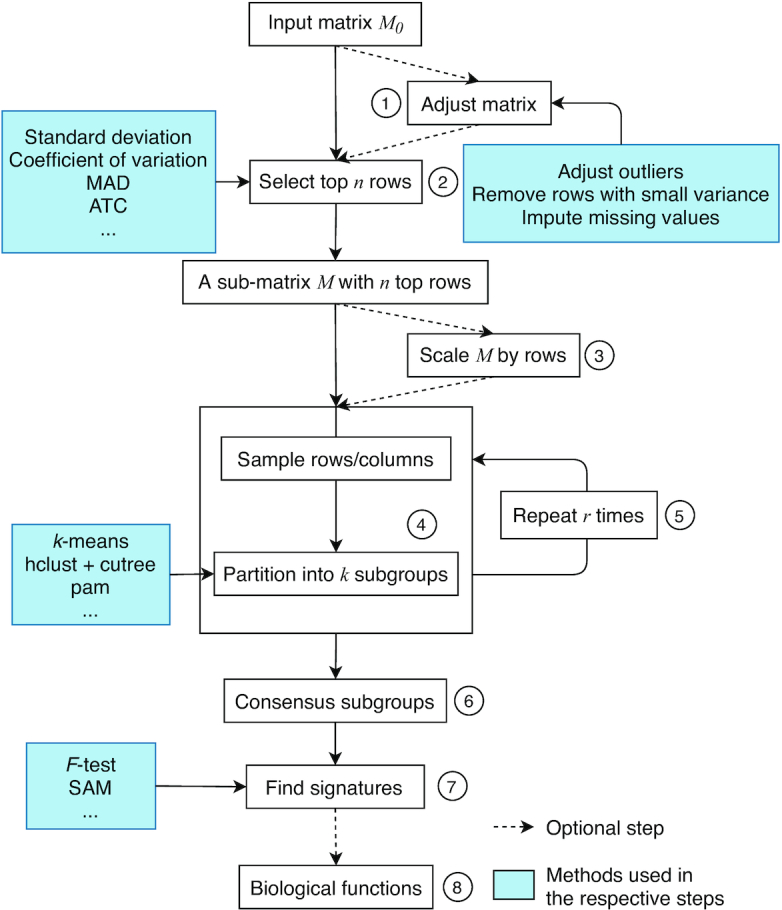
The cola workflow. Steps are as follows: 1. Adjust the input matrix by removing outliers and imputing missing values; 2. Select top n features by a certain method, e.g., standard deviation; 3. Scale the matrix by rows; 4. Randomly sample from the matrix and partition samples by a certain partitioning method; 5. Repeat Step 4 r times to obtain a list of partitions; 6. Construct the consensus partition; 7. Identify features discriminating between subgroups; 8. Apply functional enrichment on signatures if they can be annotated to genes.
