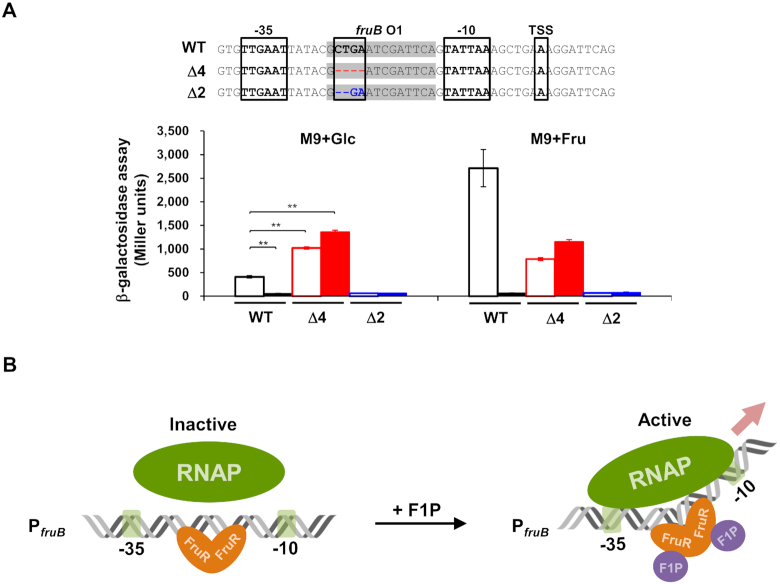
Figure 5.
Optimization of the spacer length between –35 and –10 promoter elements increased the fruB promoter activity independently of the presence of VcFruR. (A) The effect of the spacer length between –35 and –10 elements of the fruB promoter on its activity was measured by the lacZ reporter assay using a V. cholerae N16961 lacZ−fruR− (filled bars) or lacZ−fruR+ (open bars) strain harboring plasmids carrying E. coli lacZ transcriptionally fused with the modified V. cholerae fruB promoter with 2 or 4 bp deletion in the fruB O1 sequence (Δ2 and Δ4, marked as blue and red, respectively) as well as the wild-type fruB promoter (WT). Nucleotide sequences corresponding to fruB O1 are shaded in grey. The –35 and –10 promoter elements and TSS are boxed. Deleted nucleotides in the modified promoters are shown in blue and red, respectively, and marked as ‘-’ (upper panel). Indicated strains were grown on glucose or fructose and then lysed to measure the β-galactosidase activity (lower panel). Statistical significance was determined using the Student's t-test (**P< 0.01). (B) A schematic view of the proposed mechanism of transcriptional activation of the fru operon by FruR in V. cholerae. In the absence of F1P, FruR binds to the binding site fruB O1 located between –27 to –12 relative to the TSS in the fruB promoter, which has a suboptimal spacer length of 20 bp, and interferes with the binding of RNAP to the promoter. Accordingly, transcriptional activation of the fru operon does not occur. In the presence of fructose, however, it is transported through the PTSFru and concomitantly phosphorylated to F1P. Then, the FruR–F1P complex binds to fruB O1 and induces a structural change in the DNA spacer region between the -35 and -10 elements, thereby facilitating RNAP binding to the promoter to trigger the transcriptional activation of the fru operon.