Figure 4.
PRIME reveals molecular determinants of TCR recognition
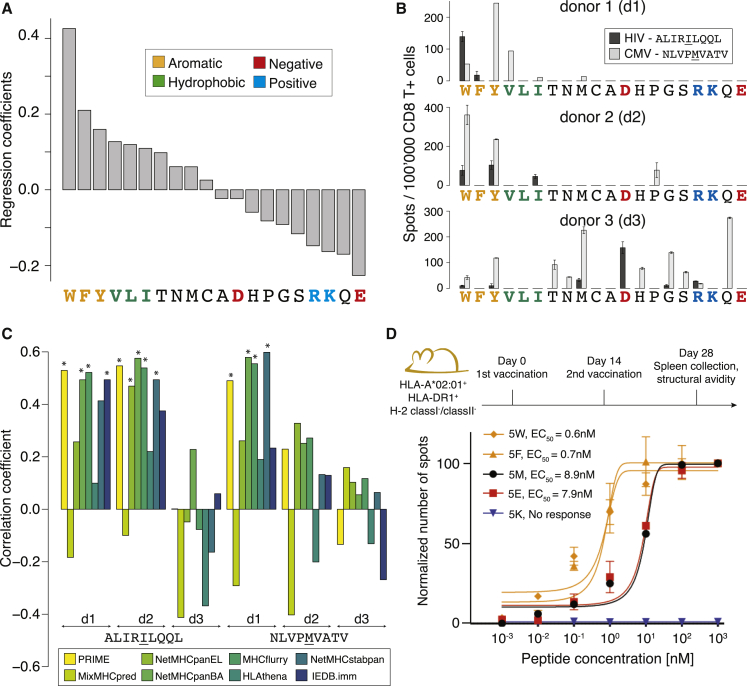
(A) Ranking of the coefficients in PRIME corresponding to amino acids at MIA positions.
(B) IFNγ-ELISpot signals obtained after stimulating naive CD8+ T cells from three healthy donors (d1, d2, and d3) with all P5 variants of the HIV (ALIRILQQL) and CMV (NLVPMVATV) epitopes (two technical replicates).
(C) Spearman correlation coefficients between immunogenicity predictions and IFNγ ELISpot signals for P5 variants of the HIV and CMV epitopes from each donor. Stars indicate Spearman correlation p values smaller than 0.05.
(D) Functional avidity of CD8+ T cell responses (EC50) measured for five variants of the CMV epitopes used to vaccinate HLA-A∗02:01 transgenic mice (4–7 biological replicates). Data were renormalized for each epitope based on the signal at 103 nM. No response was detected for the peptide with K at P5 (values of 0 are shown).