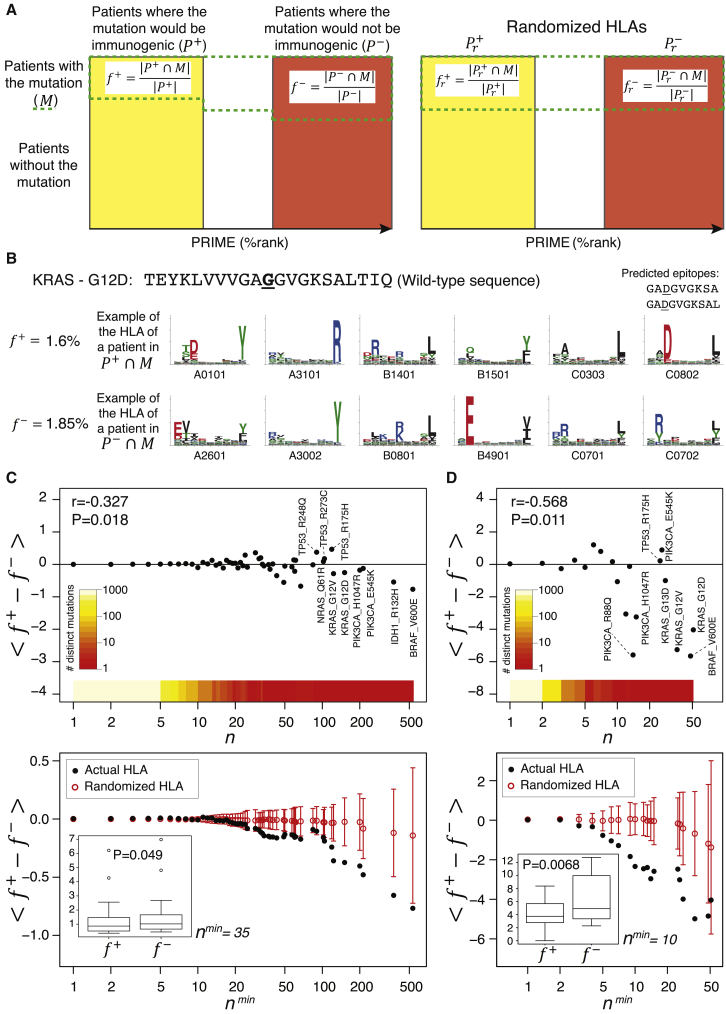
Figure 6.
PRIME provides insight into immunoediting in human cancer
(A) Proposed framework to study immunoediting acting on cancer mutations. For each mutation, patients are stratified based on whether the mutation would be immunogenic (, yellow rectangle) or would not (, red rectangle). The actual frequency of the mutation is compared between these two groups ( and , where stands for the subset of patients where the mutation is observed). These frequencies will be further compared to those obtained after shuffling the HLA-I alleles between patients (i.e., and ).
(B) Frequency of KRAS G12D mutation in patients where it would give rise to neo-epitopes () and where it would not (), together with the example of HLA-I alleles of a patient with the mutation in (upper row, with the two predicted epitopes) and one in (lower row).
(C) Top: average value of for mutations observed N times ( in the TCGA cohort. The color scale shows the number of distinct mutations for all values of the mutation occurrence (N > 0). Bottom: average value of for mutations observed at least Nmin times in the TCGA cohort is shown. The red circles and error bars correspond to randomized HLA-I alleles (mean and standard deviation of ).
(D) Top: average value of for mutations observed N times in colorectal tumors of TCGA. Bottom: average value of for mutations observed at least Nmin times in colorectal tumors of TCGA is shown.
The insets in (C) and (D) show the boxplots of the frequencies and for all mutations observed at least Nmin times in our TCGA samples (Nmin = 35 in C and Nmin = 10 in D). r and p values shown in the top plots in (C) and (D) correspond to the Pearson correlation coefficient (STAR methods). p values shown in the insets of the bottom plots in (C) and (D) correspond to paired Wilcoxon tests. All frequencies are shown in percentages (%).