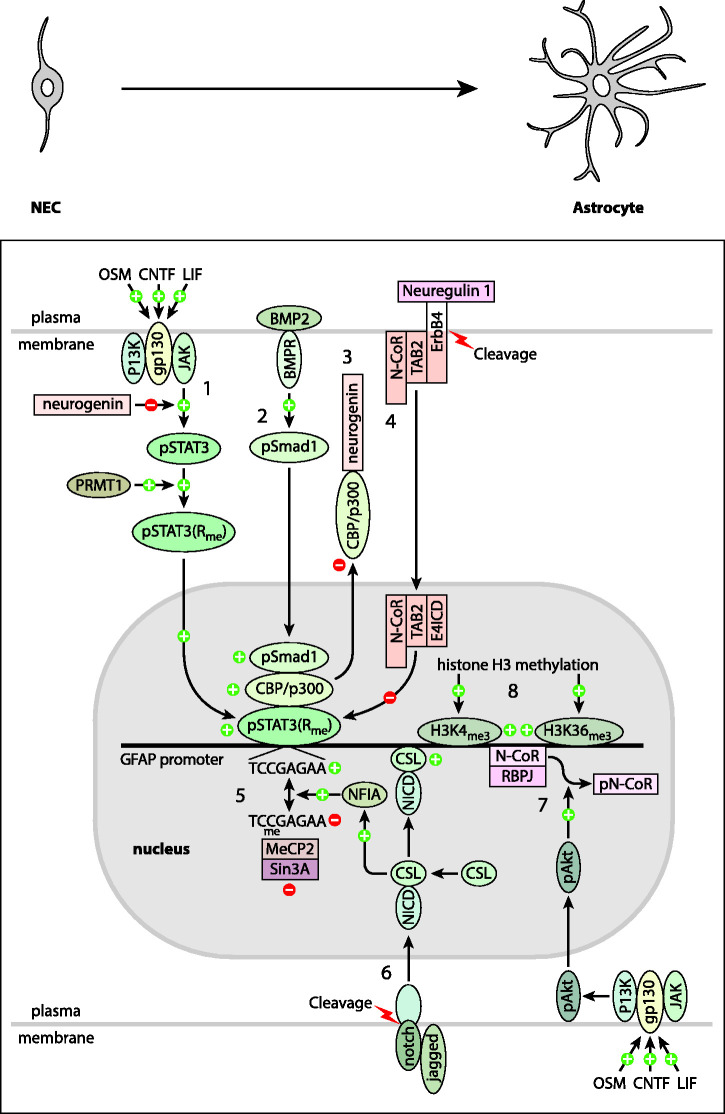
Figure 2.
Developmental Regulation of GFAP Expression. Shown are mechanisms proposed to regulate the developmental timing of GFAP expression that are discussed in the text. Plus signs (+), and ovoid shapes indicate processes that stimulate GFAP expression, whereas minus signs (–) and rectangular boxes indicate those that are inhibitory. Numbers in the figure are keyed both to this legend and the subdivisions of the section Developmental Regulation of GFAP Expression. Foremost among the positive factors is STAT3 [1]. It is activated by cytokines such as CNTF, LIF and oncostatin M (OSM) binding to their receptors complexed with gp130, resulting in the activation of Janus kinase (JAK). JAK then activates STAT3 by phosphorylation. pSTAT is further activated by methylation by protein arginine methyltransferase 1 (PRMT1) (it is not known if this occurs in the cytoplasm or nucleus). pSTAT3(Rme) (hereafter STAT3) enters the nucleus and attaches to its consensus binding sequence, TCCGAGAA, in the GFAP promoter. Assisting STAT3 in its stimulation of GFAP expression is Smad1, which is activated following binding of BMP2 to its plasma membrane receptor [2]. Smad1 is believed to form a complex with STAT3 via mutual interaction with CBP/p300. Several mechanisms prevent premature expression of GFAP, many targeting the activity of STAT3. Neurogenin, which is present at high levels early in development, inhibits the phosphorylation of STAT3 by JAK, and also suppresses GFAP expression by sequestering CBP/p300 [3]. STAT3 activity is also inhibited by a complex of TAB2, N-CoR, and the intracellular domain of the receptor ErbB4 (E4ICD), which is generated by cleavage of ErbB4 following neuregulin 1 binding [4]. In addition, binding of STAT3 to its consensus sequence is inhibited by methylation of the CpG within the consensus sequence [5]. The CpG methylation also contributes to preventing GFAP expression by attracting the transcriptional repressor Sin3A via its binding to MeCP2. Another inhibitory mechanism is binding of the transcriptional repression complex RBPJ/N-CoR to the GFAP promoter [7]. Inhibition is relieved by developmental decreases in the levels of Sin3A, neurogenin and neuregulin. Notch signaling also contributes to activation of GFAP expression through generation of the Notch intracellular domain (NICD) by proteolytic cleavage following binding of a ligand such as jagged [6]. The NICD then forms a complex with CSL, a transcriptional activator, which binds to the GFAP promoter. In addition, by unknown mechanisms, the NICD/CSL complex increases levels of NFIA, which facilitates demethylation of the CpG in the STAT3 binding site. Also contributing to GFAP expression is activation of Akt by phosphorylation via the PI3K pathway (a single arrow is shown, but multiple steps are involved) [7]. pAkt in turn phosphorylates the transcriptional repressor N-CoR, causing it to exit the nucleus. Developmental increases in methylation of lysines 4 and 36 of histone 3 are also associated with increased GFAP transcription [8]. A stimulatory mechanism not illustrated, but discussed in the text in the section BRG1 and Gene Clustering, is association of the GFAP promoter with other STAT-activated genes via the bridging protein BRG1.