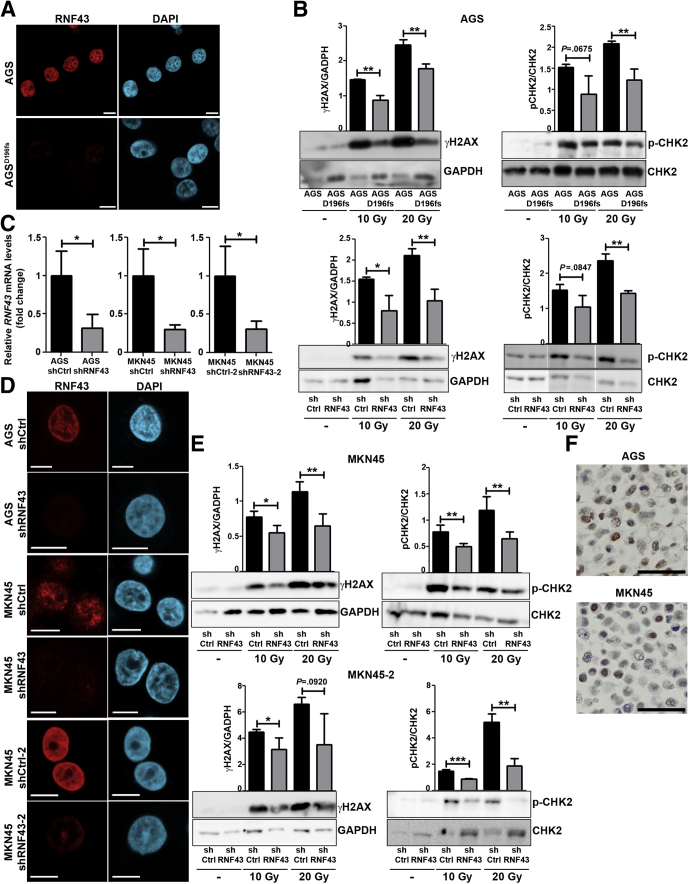
Figure 1.
RNF43 is involved in DDR. (A) Representative immunofluorescence images of RNF43 (red) in control and AGSD196fs cells. Scale bars: 10 μm. (B) Western blot analysis and quantification of γH2AX and CHK2 expression in WT AGS, AGSD196fs, control AGS (shCtrl), and RNF43 knockdown AGS (shRNF43) cells after γ-radiation. GAPDH was used as a protein loading control (N = 3). (C) RNF43 mRNA levels in control and RNF43 knockdown AGS and MKN45 cells. Cycle threshold (CT)values were normalized to GAPDH and fold change was calculated over control cells. (D) Representative immunofluorescence images of RNF43 (red) in control and RNF43 knockdown AGS and MKN45 cells. Scale bars: 10 μm. (E) Western blot analysis and quantification of γH2AX and CHK2 expression in control MKN45 (shCtrl) and RNF43 knockdown MKN45 (shRNF43) cells after γ-radiation. GAPDH was used as a protein loading control (N = 3). (F) Endogenous RNF43 expression in AGS and MKN45 cells detected by immunocytochemistry. Scale bar: 50 μm. Error bars indicate SD. ∗P ≤ .05, ∗∗P ≤ .01, ∗∗∗P ≤ .001, 2-tailed unpaired t test. DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; mRNA, messenger RNA.