Fig. 6.
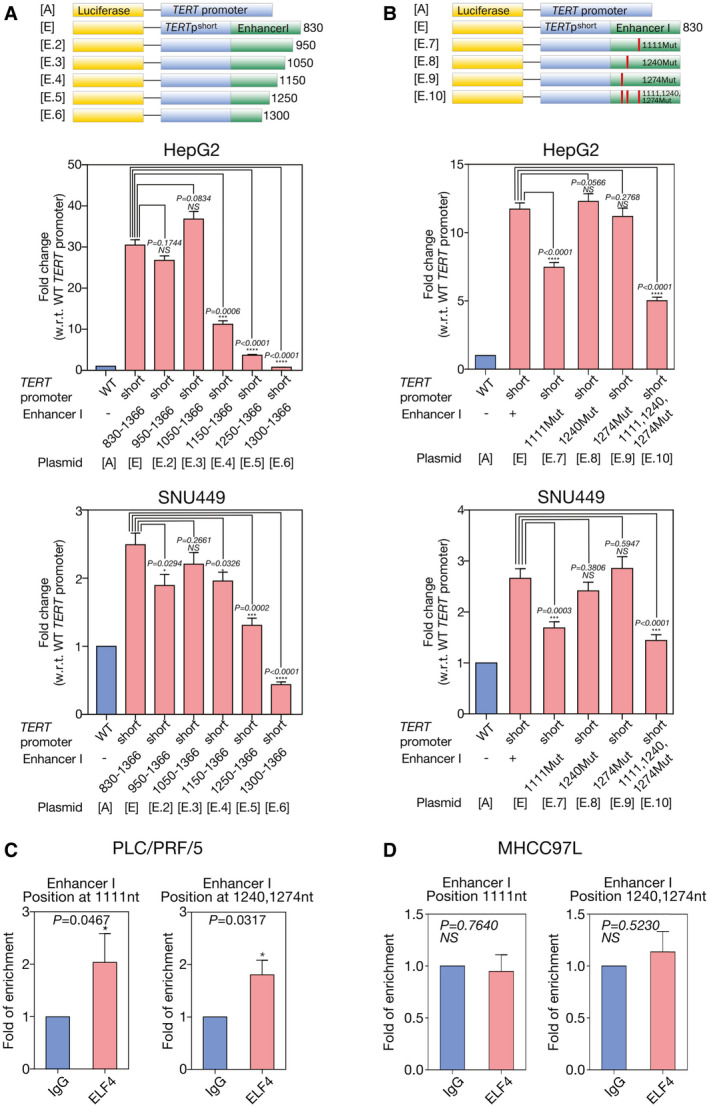
Binding of ELF4 at HBV EnhI region resulted in enhanced TERT HBV chimeric fusion promoter activity. (A) The promoter activity of TERT HBV chimeric fusion promoter was significantly reduced on deletion of the 830‐1150 HBV EnhI region (TERTpshortHBx‐EnhI(Δ830‐1150), plasmid E.4; 63.2% reduction, P < 0.0001), 830‐1250 region (TERTpshortHBx‐EnhI(Δ830‐1250), plasmid E.5; 87.9% reduction, P < 0.0001), and 830‐1300 region (TERTpshortHBx‐EnhI(Δ830‐1300), plasmid E.6; 97.5% reduction, P < 0.0001) but not for deletion of the 830‐950 (TERTpshortHBx‐EnhI(Δ830‐950), plasmid E.2) and 830‐1050 (TERTpshortHBx‐EnhI(Δ830‐1050), plasmid E.3) regions (both P > 0.05) in HepG2 cells. In SNU449 cells, the promoter activity of TERT HBV chimeric fusion promoter was significantly reduced on deletion of the 830‐1250 region (TERTpshortHBx‐EnhI(Δ830‐1250), plasmid E.5; 47.4% reduction, P = 0.0002) and 830‐1300 region (TERTpshortHBx‐EnhI(Δ830‐1300), plasmid E.6; 82.4% reduction, P < 0.0001). (B) The promoter activity of TERT HBV chimeric fusion promoter was significantly reduced on single mutation of the putative ELF4 binding site at position 1,111 (TERTpshortHBx‐EnhI+(1111Mut), plasmid E.7; 36.2% reduction, P < 0.0001) and triple putative ELF4 binding site mutations at positions 1,111, 1,240, and 1,274 (TERTpshortHBx‐EnhI+(1111,1240,1274Mut), plasmid E.10; 57.3% reduction, P < 0.0001) although not on mutating single putative ELF4 binding sites at positions 1,240 (TERTpshortHBx‐EnhI+(1240Mut), plasmid E.8) and 1,274 (TERTpshortHBx‐EnhI+(1274Mut), plasmid E.9) of the HBV DNA region as compared with the wild‐type TERT HBV chimeric fusion (TERT shortHBx‐EnhI+, plasmid E) promoter in HepG2 cells. In SNU449 cells, the promoter activity of TERT HBV chimeric fusion promoter was significantly reduced on single mutation of the putative ELF4 binding site at position 1,111 (TERTpshortHBx‐EnhI+(1111Mut), plasmid E.7; 36.6% reduction, P < 0.0001) and triple putative ELF4 binding site mutations at positions 1,111, 1,240, and 1,274 (TERTpshortHBx‐EnhI+(1111,1240,1274Mut), plasmid E.10; 45.8% reduction, P < 0.0001). (C) ChIP assay using primers flanking positions 1,000‐1,120 and 1,240‐1,300 of HBV DNA region showed ELF4 physically interacted with HBV EnhI region in PLC/PRF/5. (D) There was no significant interaction of ELF4 with HBV EnhI region at positions between 1,000‐1,120 and 1,240‐1,300 in MHCC‐97L on ChIP assay. ***P < 0.001; ****P < 0.0001. Mut, mutation; NS, not significant; w.r.t., with regard to; WT, wild type.
