Figure 3.
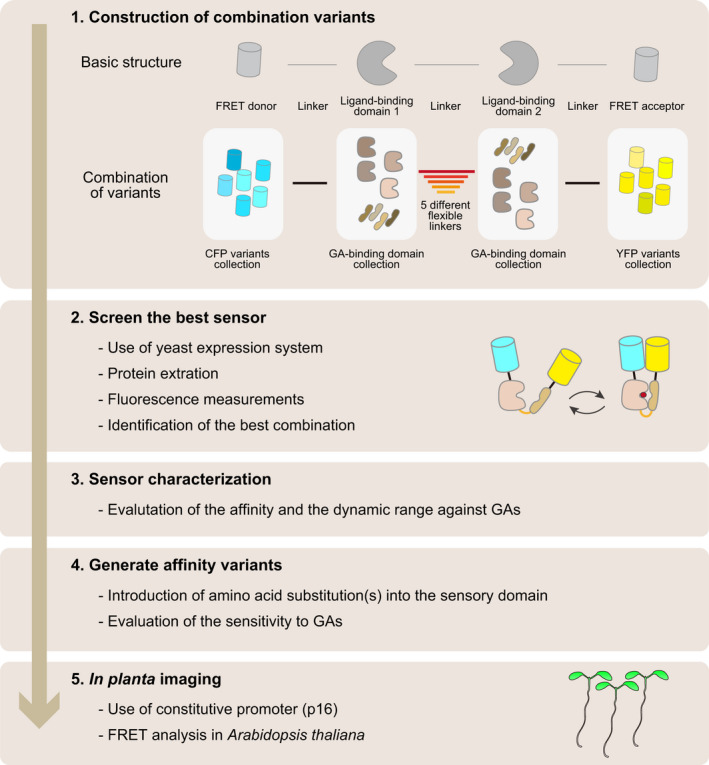
Flow chart for biosensor engineering. We present the engineering procedure for a plant hormone biosensor, in this instance the gibberellin (GA) sensor GPS1 (Jones et al., 2014; Rizza et al., 2017). The biosensor was created by combining domains of different bipartite GA receptors through an artificial linker and flanking the ligand‐binding domains with potential Förster resonance energy transfer (FRET) pairs. Linkers and collections of fluorescent protein (FP) variants were tested as FRET pairs and compared in high‐throughput yeast expression assays. Candidate GA‐binding domains were selected from the receptors AtGID1A, AtGID1B and AtGID1C, and truncated AtGAI or AtRGA domains (DELLA protein domain). The GA‐binding domains were linked to each other by different linkers. The combinatorial library was constructed using Gateway® cloning technology, introduced into a protease‐deficient yeast strain, expressed and isolated. The best five‐way combination was screened by in vitro fluorescence measurements of the sensors isolated from yeast. Affinity mutants with reduced or no response to GAs were engineered by rational design. The affinity mutants can be used as negative controls during in vivo analysis to exclude artifacts such as those caused by pH changes. GPS1 and the affinity mutant GPS1‐NR were expressed in Arabidopsis plants under the control of a constitutive promoter (p16).
