FIGURE 7.
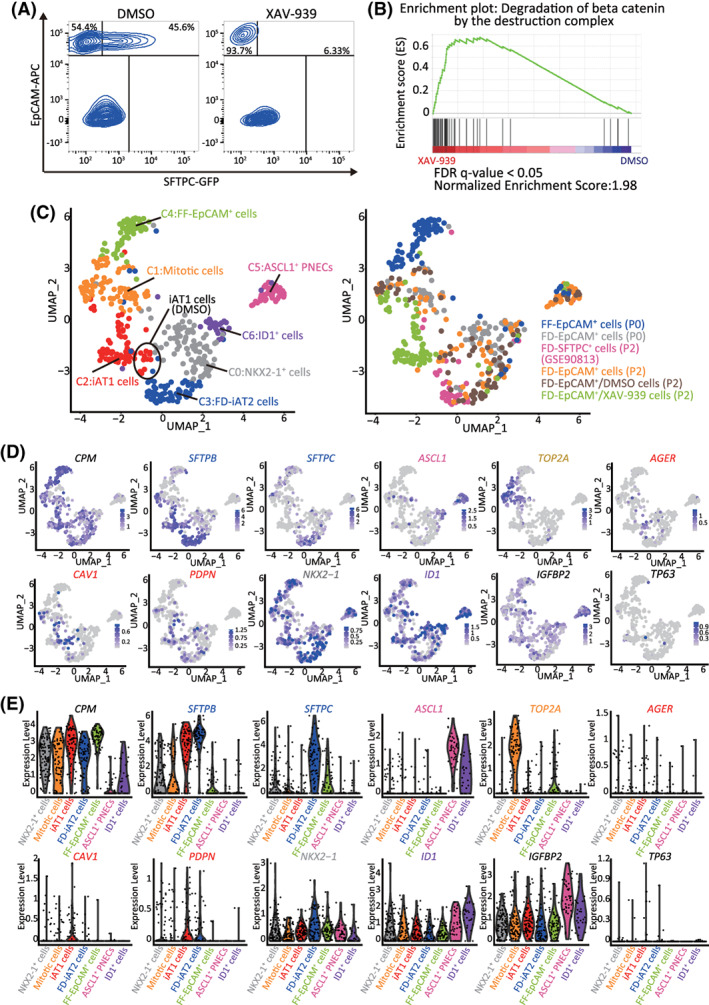
XAV‐939 promotes iAT1 cell differentiation in FD‐AOs. A, FD‐AOs (P2) were treated with 10 μM XAV‐939 or DMSO (vehicle control) from day 2 to 14, and then the FD‐EpCAM+ cells collected using FACS were subjected to scRNA‐seq. B, GSEA using the gene set of β‐catenin degradation by the destruction complex. Significantly up‐ or downregulated genes (P < .05) ranked by the log difference in average gene expression between DMSO‐ and XAV‐939‐treated FD‐EpCAM+ cells (P2) were used for the analysis. C, UMAP analysis showing cell clusters in XAV‐939‐treated FD‐AOs. XAV‐939‐treated FD‐EpCAM cells (P2) were included among iAT1 cells. D,E, UMAP and violin plots showing the expression of the representative lung epithelial cell marker genes; AT1 cells (AGER, CAV1, and PDPN), AT2 cells (SFTPB and SFTPC), mitotic cells (TOP2A), ASCL1+ PNECs (ASCL1), ID1+ cells (ID1), NKX2‐1+ cells (NKX2‐1), basal cells (TP63), subpopulation of AT1 cells (IGFBP2), and alveolar epithelial cells (CPM). DMSO, dimethyl sulfoxide; FACS, fluorescence‐activated cell sorting; FD‐AOs, fibroblast‐dependent alveolar organoids; GSEA, gene set enrichment analysis; scRNA‐seq, single‐cell RNA sequencing; UMAP, uniform manifold approximation and projection
