Figure 1.
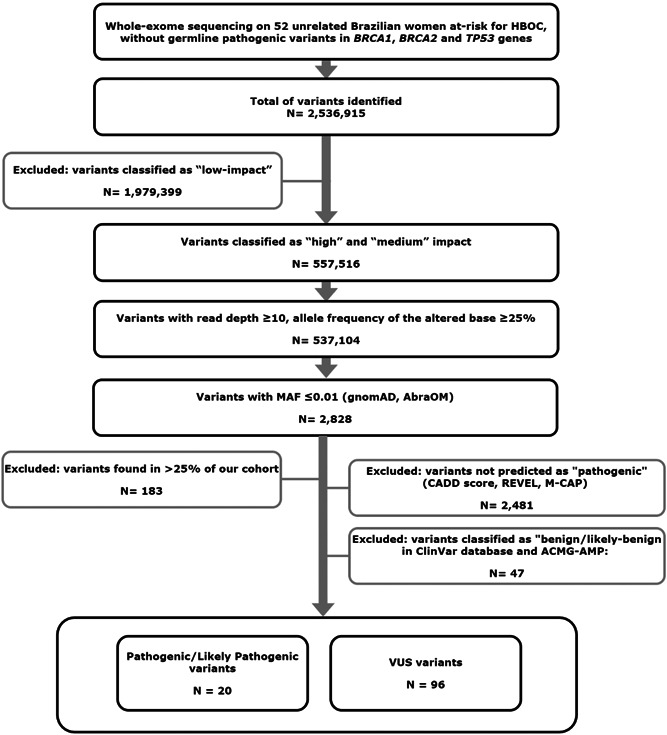
Variants selection workflow. Whole‐exome sequencing data from 52 unrelated Brazilian women at‐risk for HBOC, without germline pathogenic variants in BRCA1, BRCA2, and TP53 genes. Variants classified as “high‐impact” and “medium‐impact” by snpEFF/GEMINI were prioritized. Then, variants with base coverage more than or equal to 10× and variant allele frequency (VAF) more than or equal to 0.25 were selected, and those present in population databases with frequency less than or equal to 1% (MAF ≤ 0.01) were analyzed. The variants were also separated accordingly to ClinVar and ACMG classification. HBOC, hereditary breast and ovarian cancer
