Figure 2.
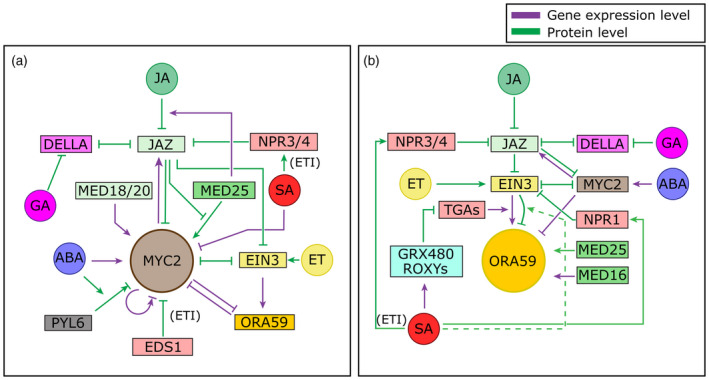
Schematic overview of hormone crosstalk acting on two key transcription factors, MYC2 and ORA59, of the two branches of the jasmonic acid (JA) pathway.
(a) Crosstalk acting on the MYC branch master regulator MYC2. In the context of defense, MYC2 mostly regulates anti‐insect responses. MYC2 is repressed by interacting JAZ repressors, and MYC2 itself can induce transcription of these JAZs. JA induces the breakdown of JAZs, thus leading to activation of MYC2. MED25 promotes MYC2 transcriptional activity, but JAZs prevent binding of MED25 to MYC2. MED25 also promotes JAZ breakdown by recruiting COI1, and alters JAZ splicing and thereby JAZ sensitivity to JA. MED18 and MED20 promote transcription of MYC2. EIN3 is activated by JA‐mediated breakdown of JAZ proteins and ethylene (ET)‐mediated stabilization. It binds to and represses MYC2 and vice versa. EIN3 also transcriptionally activates ORA59 and ORA59 can repress MYC2 transcription and vice versa (either directly or indirectly, see also panel b). MYC2 can enhance its own transcription in the short term but represses it in the long term. During AvrRps4‐induced ETI, EDS1 can repress MYC2. Furthermore, SA can promote degradation of JAZs via NPR3 and NPR4 during ETI. Generally, SA is an inhibitor of MYC2 transcription. Abscisic acid (ABA) directly activates transcription of MYC2 and enhances binding of the ABA receptor PYL6 to MYC2, modulating transcriptional activity of MYC2, which differentially acts on the JAZ6 and JAZ8 promoters (leading to repression versus activation). DELLA proteins bind JAZs and thereby JAZs and DELLAs prevent each other from binding to their respective target transcription factors (TFs). Gibberellin (GA) induces breakdown of DELLAs and thus indirectly represses MYC2. (b) Crosstalk acting on the ERF branch master regulator ORA59. ORA59 mostly regulates defense against necrotrophic pathogens. ORA59 is indirectly regulated by both JA and ET through their action on EIN3: JA releases EIN3 from its repression by JAZ and ET stabilizes EIN3. When released from repression and degradation, EIN3 activates transcription of ORA59. TGA TFs are also needed for this activation. GRX480 and other ROXYs are induced by SA and repress the transcriptional activity of these TGAs, leading to reduced transcription of ORA59. EIN3 also mediates degradation of ORA59. Because this only leads to reduced ORA59 functioning under high SA levels and not under high ET levels we propose that SA specifically modulates EIN3 activity such that it leads to ORA59 degradation (dotted line). SA activates NPR1’s activity as a co‐transcriptional regulator. NPR1 can interact with EIN3, leading to repression of EIN3 transcriptional activity. We propose that it is unlikely that NPR1 modulates EIN3‐mediated ORA59 activation during SA/JA crosstalk (see the section ‘Crosstalk by modulation of protein stability’). EIN3 is further repressed by MYC2 through direct binding and this also occurs the other way around. MYC2 is repressed by interaction of JAZ repressors and itself activates transcription of these JAZs. It is also transcriptionally activated by ABA. ORA59 expression is inhibited by MYC2, both directly and possibly indirectly via inhibition of EIN3 by MYC2. DELLAs bind to JAZs and thereby they inhibit each other from binding to target TFs in their respective pathways. GA leads to breakdown of DELLAs, thus indirectly repressing ORA59. MED25 interacts with ORA59 and promotes its transcriptional activity, and MED16 promotes ORA59 transcription. Modulation of JAZ breakdown and JAZ RNA splicing by MED25 is not shown here (see panel a). During ETI SA can promote degradation of JAZs via NPR3 and NPR4. Note that in most cases where EIN3 is mentioned, EIL1 likely has the same function. However, in most research only EIN3 is extensively characterized.
Mechanisms acting on the gene expression level are colored purple and mechanisms acting on the protein level are colored green. Arrows and bar‐headed lines indicate positive and negative effects, respectively. Mechanisms acting downstream of MYC2 and ORA59 or at the hormone homeostasis level are not shown.
