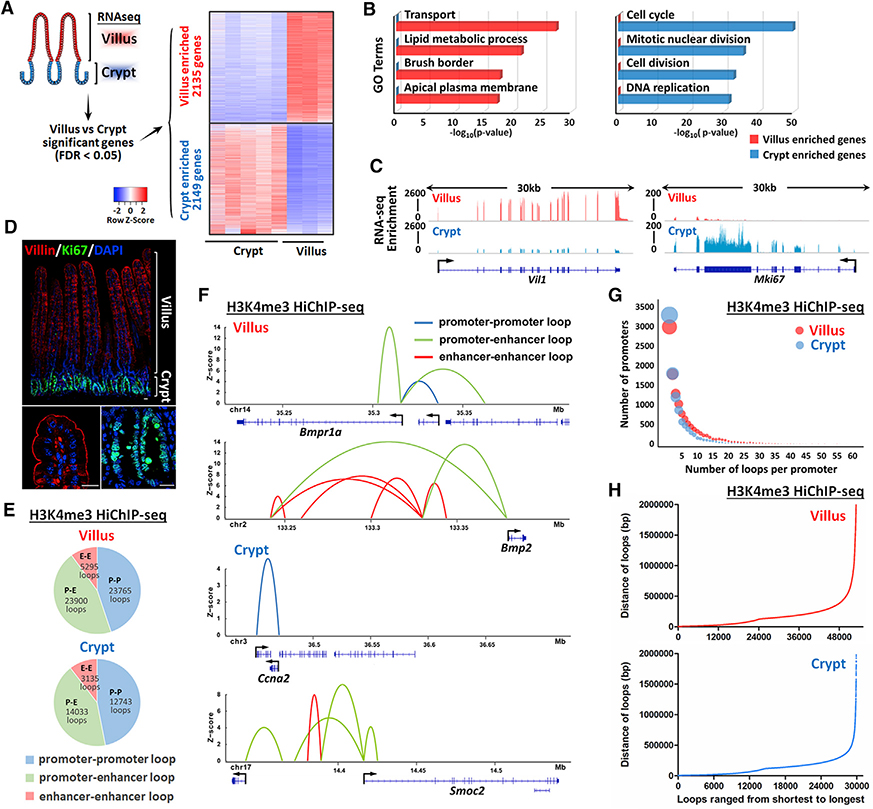
Figure 1. H3K4me3 HiChIP catalogs 3D chromatin looping events from promoters of the intestinal epithelium.
(A) Heatmap of villus-enriched and crypt-enriched genes identified by RNA-seq analysis reveals a dynamic transcriptome as cells transition from crypts and differentiate on villi (n = 5 crypts and 3 villi; Cuffdiff FDR < 0.05; GEO: GSE53545, GSE70766, and GSE102171).
(B) Functional annotation (DAVID) of villus-enriched and crypt-enriched genes. p values were calculated using DAVID (see full table in Table S1).
(C and D) Examples are shown by (C) RNA-seq tracks and (D) immunofluorescence staining that Villin and Ki67 are enriched in villus and crypt compartment, respectively.
(E–H) H3K4me3 HiChIP-seq reveals chromatin interactions of isolated duodenal crypt or villus epithelium (n = 2 biological replicates). Genomic regions harboring significant interactions (q ≤ 0.0001 and counts ≥ 4, 2 replicates each condition) were analyzed for their chromatin profiles across the crypt-villus axis. Loops with q ≤ 0.0001 and counts ≥ 8 (combined 2 replicates) were visualized with the Sushi package.
(E and F) Percentage (E) and examples (F) of promoter-promoter loops, promoter-enhancer loops, and enhancer-enhancer loops identified in villus and crypt cells.
(G) Number of significant chromatin interactions identified for each annotated promoter (UCSC transcription start site). The diameter of each circle corresponds to the number of promoters.
(H) Distribution in the distance of significant chromatin loops identified by H3K4me3 HiChIP-seq.