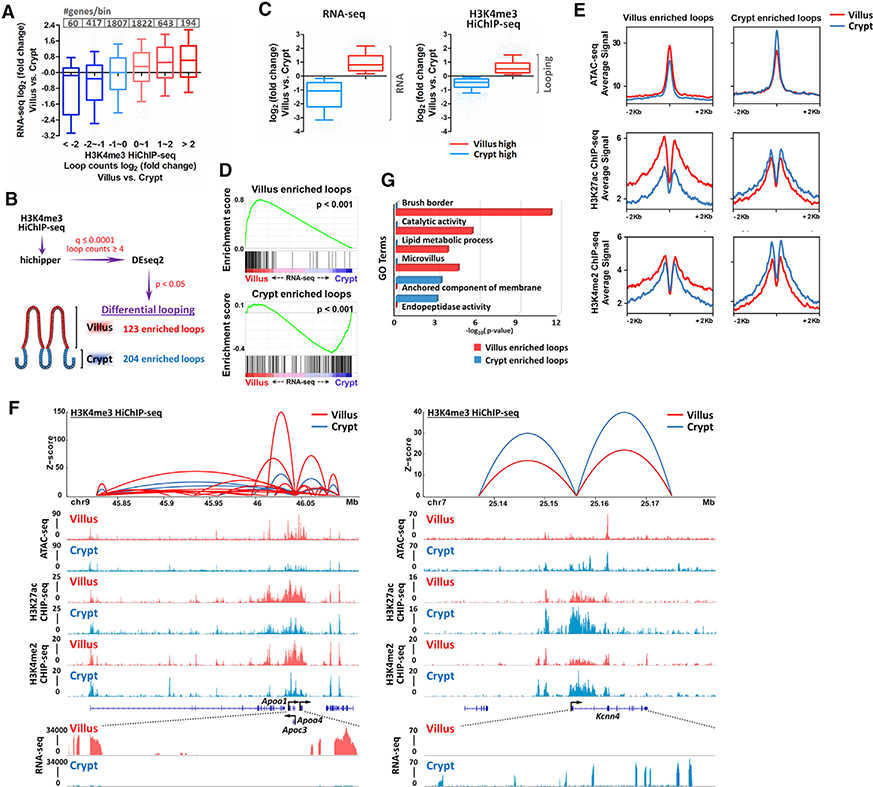
Figure 2. Differential promoter looping across the crypt-villus axis.
(A) Promoter looping events are correlated with RNA transcript levels of their nearby genes (within 1 kb of transcriptional start sites [TSSs]).
(B) Schematic of differential looping events identified by H3K4me3 HiChIP-seq between villus and crypt samples.
(C) Boxplots show the magnitude of the changes in RNA transcript levels or looping strengths, as measured by RNA-seq FPKM counts of genes detected by Cuffdiff analysis, or H3K4me3 HiChIP-seq read counts (q ≤ 0.0001 and counts ≥ 4), respectively. Boxplot line represents the median; whiskers represent the 10th and 90th percentile (Mann-Whitney test, p < 0.001).
(D–F) Transcript levels of linked genes and levels of active chromatin marks are positively correlated with villus- and crypt-enriched looping events (differential loops, DESeq2 p < 0.05).
(D) Gene set enrichment analysis reveals that genes nearby villus-enriched loops are highly expressed in villus cells, whereas nearby genes of crypt-enriched loops are highly expressed in crypt cells (within 10 kb of TSSs; Kolmogorov-Smirnov test, p < 0.001).
(E) SitePro plots show the average signal profiles of active chromatin markers around villus and crypt-enriched looping regions.
(F) Integrated datasets are shown as examples for HiChIP (combined replicates with loop counts ≥ 8 and q ≤ 0.0001), ATAC-seq, ChIP-seq, and RNA-seq at the indicated villus- and crypt-enriched looping regions.
(G) Functional annotation (DAVID) of genes nearby (within 10 kb of TSSs) villus-enriched and crypt-enriched loops.
p values were calculated using DAVID (see full table in Table S3). RNA-seq (GEO: GSE53545, GSE70766, and GSE102171): n = 5 crypts and 3 villi; H3K4me3 HiChIP-seq: n = 2 biological replicates; ATAC-seq: n = 3 biological replicates; ChIP-seq: one replicate was used for each active chromatin mark, and two active chromatin marks were tested in total.