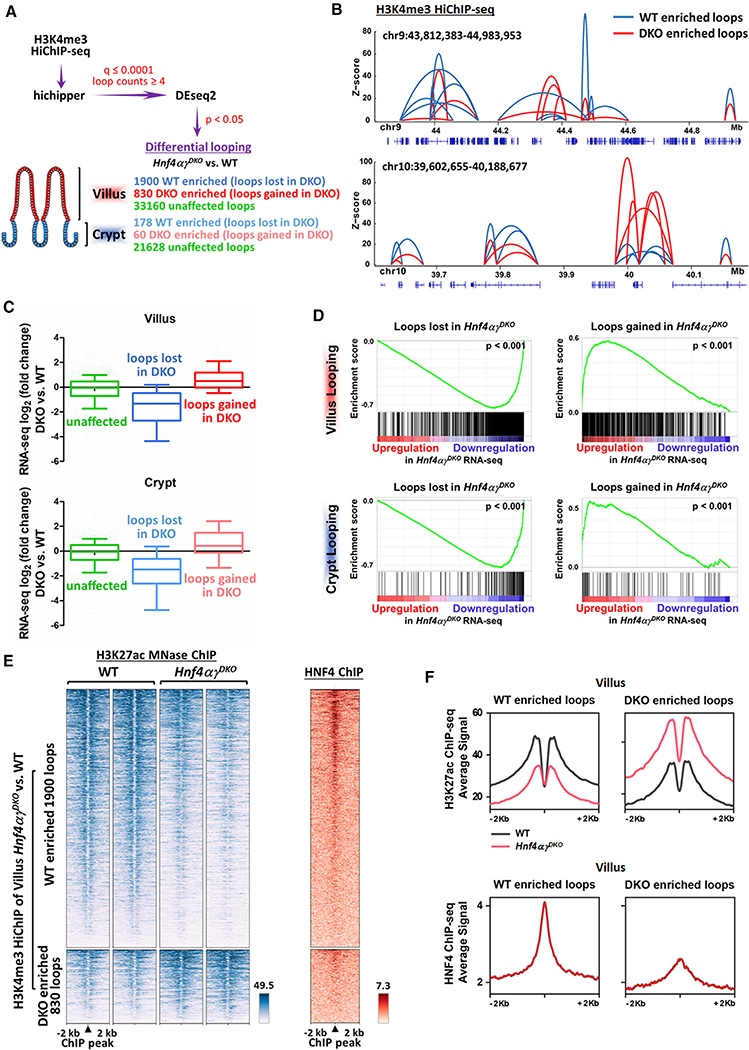
Figure 3. HNF4 transcription factors are required for chromatin looping.
(A) Schematic of differential loop calling by H3K4me3 HiChIP-seq between WT and Hnf4αγDKO samples.
(B) Examples of WT- and Hnf4αγDKO-enriched loops (differential loops, DESeq2 p < 0.05).
(C and D) Boxplots (post hoc Dunn’s test was applied following a Kruskal-Wallis test, p < 0.001; C) and GSEA (Kolmogorov-Smirnov test, p < 0.001; D) show that transcriptome level changes in Hnf4αγDKO versus WT (TSS distances in 10-kb windows) are correlated with WT-enriched (loops lost in Hnf4αγDKO) and Hnf4αγDKO-enriched (loops gained in Hnf4αγDKO) looping events from the gene’s promoter. Boxplot line represents the median; whiskers represent the 10th and 90th percentile.
(E and F) Heatmap (E) and SitePro (F) plots show that WT-enriched and Hnf4αγDKO-enriched looping events are correlated with changes in H3K27ac signal of Hnf4αγDKO versus WT and HNF4 binding events. H3K4me3 HiChIP-seq: n = 2 biological replicates; RNA-seq (WT versus Hnf4αγDKO; GSE112946): n = 3 biological replicates; H3K27ac ChIP-seq (WT versus Hnf4αγDKO; GSE112946): n = 2 biological replicates; HNF4 ChIP-seq (WT versus Hnf4αγDKO; GSE112946): n = 2 biological replicates for each HNF4 paralog.