Abstract
Spillback transmission from humans to animals, and secondary spillover from animal hosts back into humans, have now been documented for SARS-CoV-2. In addition to threatening animal health, virus variants arising from novel animal hosts have the potential to undermine global COVID-19 mitigation efforts. Numerous studies have therefore investigated the zoonotic capacity of various animal species for SARS-CoV-2, including predicting both species’ susceptibility to infection and their capacities for onward transmission. A major bottleneck to these studies is the limited number of sequences for ACE2, a key cellular receptor in chordates that is required for viral cell entry. Here, we combined protein structure modeling with machine learning of species’ traits to predict zoonotic capacity of SARS-CoV-2 across 5,400 mammals. High accuracy model predictions were strongly corroborated by in vivo empirical studies, and identify numerous mammal species across global COVID-19 hotspots that should be prioritized for surveillance and experimental validation.
Keywords: coronavirus, COVID-19, hosts, reservoirs, zoonotic, spillover, spillback, susceptibility, machine learning, homology modelling, ACE2
Introduction
The ongoing COVID-19 pandemic has surpassed 2.4 million deaths globally as of 17 February 2021 (Dong et al., 2020; WHO, 2021). Like previous pandemics in recorded history, COVID-19 originated from the spillover of a zoonotic pathogen, SARS-CoV-2, a betacoronavirus originating from an unknown animal host (Gage and Kosoy, 2005; Keele et al., 2006; Taubenberger et al., 2005; P. Zhou et al., 2020). The broad host range of SARS-CoV-2 is due in part to its use of a highly conserved cell surface receptor to enter host cells, the angiotensin-converting enzyme 2 receptor (ACE2) (Letko et al., 2020). This receptor is found in all major vertebrate groups (Chou et al., 2006).
The ubiquity of ACE2 coupled with the high prevalence of SARS-CoV-2 in the global human population explains multiple observed spillback infections in the past year. In spillback infection, human hosts transmit SARS-CoV-2 virus to cause infection in non-human animals. In addition to threatening wildlife and domestic animals, repeated spillback infections may lead to the establishment of new animal hosts from which SARS-CoV-2 can continue to pose a risk of secondary spillover infection to humans through bridge hosts (e.g., (Guth et al., 2019) or newly established enzootic reservoirs. Indeed, this risk has already been realized in Denmark (WHO, 2020) and The Netherlands, where SARS-CoV-2 spilled back from humans to farmed mink (Neovison vison) and a variant of SARS-CoV-2 was subsequently transmitted from mink back to humans (Oude Munnink et al., 2020). This exemplifies a major concern in these secondary spillover events, where a mutant strain arising somewhere along the transmission chain (Garry, 2021; Oude Munnink et al., 2020) affects host range (Rodrigues et al., 2020) or leads to distinct epidemiology in humans (e.g., via increased transmissibility among humans (Davies et al., 2020; Volz et al., 2021), but see (Rambaut et al., 2020; Tegally et al., 2020)). Preliminary evidence shows that the mink-derived variant exhibits moderately reduced sensitivity to neutralizing antibodies (WHO, 2020), raising concerns that humans may eventually experience more virulent infections from spillback variants, and that vaccines may eventually become less efficient at conferring immunity to variants (Van Egeren et al., 2020).
Spillback infections are already occurring worldwide. In addition to secondary spillover infections from mink farms, SARS-CoV-2 has been found for the first time in wild and escaped mink in multiple states in the United States, with viral sequences confirming that the SARS-CoV-2 variant from wild mink was identical to that found in nearby farmed mink (DeLiberto and Shriner, 2020; ODA, 2020; Shriner et al., 2021). A variety of pets, domesticated animals, zoo animals, and wildlife have also been documented as new hosts of SARS-CoV-2 (Table 1). The increasing range of known hosts for SARS-CoV-2 and the global scale of human infections signal that SARS-CoV-2 will continue to establish new enzootic infection cycles in animals, making ongoing disease control more costly and difficult. In response, recent computational studies make predictions about animal species that are most likely to be susceptible to SARS-CoV-2 (Ahmed et al., 2021; Damas et al., 2020; Huang et al., 2020; Kumar et al., 2020; S. D. Lam et al., 2020; Liu et al., 2020; Luan et al., 2020; Mathavarajah et al., 2020; Melin et al., 2020; Rodrigues et al., 2020). These studies compare sequences of ACE2 orthologs among species (sequence-based), or model the structure of the viral spike protein bound to ACE2 orthologs (structure-based) and yield a wide range of predictions about species susceptibility to SARS-CoV-2 infection. These different approaches show varying degrees of agreement with laboratory animal experiments (Figure 1).
Table 1.
Species with confirmed suitability for SARS-CoV-2 infection from natural infections or in vivo experiments. Asterisks reference species with infection status from preprints (not yet peer-reviewed). Some species (e.g, dogs) with natural infection studies also have in vivo experimental studies.
| Species | Susceptibility | Study type | Location | References |
|---|---|---|---|---|
| Cow (Bos taurus) | Yes | In vivo experiment | Lab | (Ulrich et al., 2020) |
| Dog (Canis lupus familiaris) | Yes | Natural infection | Multiple countries | (Hamer et al., 2020; OIE, 2021; Shi et al., 2020; Sit et al., 2020; USDA, 2020) |
| African green monkey (Chlorocebus aethiops) | Yes | In vivo experiment | Lab | (Woolsey et al., 2020) |
| Big brown bat (Eptesicus fuscus) | No | In vivo experiment | Lab | (Hall et al., 2020) |
| Cat (Felis catus) | Yes | Natural infection | Multiple countries | (Hamer et al., 2020; OIE, 2021; USDA, 2020; Zhang et al., 2020) |
| Gorilla (Gorilla gorilla) | Yes | Natural infection | Zoo | (San Diego Zoo, 2021) |
| Crab-eating macaque (Macaca fascicularis) | Yes | In vivo experiment | Lab | (Rockx et al., 2020) |
| Rhesus macaque (Macaca mulatta) | Yes | In vivo experiment | Lab | (Munster et al., 2020) |
| Golden hamster (Mesocricetus auratus) | Yes | In vivo experiment | Lab | (Sia et al., 2020) |
| House mouse (Mus musculus) | No | In vivo experiment | Lab | (Bao et al., 2020) |
| Ferret (Mustela putorius furo) | Yes | In vivo experiment | Lab | (Shi et al., 2020) |
| American mink (Neovison vison) | Yes | Natural infection | Multiple countries | (OIE, 2021; Oreshkova et al., 2020; USDA, 2020) |
| Raccoon dog (Nyctereutes procyonoides) | Yes | In vivo experiment | Lab | (Freuling et al., 2020) |
| European rabbit (Oryctolagus cuniculus) | Yes | In vivo experiment | Lab | (Mykytyn et al., 2021) |
| Lion (Panthera leo) | Yes | Natural infection | Multiple countries | (Bartlett et al., 2021; OIE, 2021) |
| Tiger (Panthera tigris) | Yes | Natural infection | USA and Sweden | (Bartlett et al., 2021; OIE, 2021; USDA, 2020; Wang et al., 2020) |
| Deer mouse (Peromyscus maniculatus)* | Yes | In vivo experiment | Lab | (Fagre et al., 2020; Griffin et al., 2020), |
| Cougar (Puma concolor) | Yes | Natural infection | South Africa | (OIE, 2021) |
| Egyptian fruit bat (Rousettus aegyptiacus) | Yes | In vivo experiment | Lab | (Schlottau et al., 2020) |
| Pig (Sus scrofa) | No | In vivo experiment | Lab | (Schlottau et al., 2020; Shi et al., 2020) |
| Northern treeshrew (Tupaia belangeri) | Yes | In vivo experiment | Lab | (Zhao et al., 2020) |
| Snow leopard (Uncia uncia) | Yes | Natural infection | Zoo | (Louisville Zoo, 2020) |
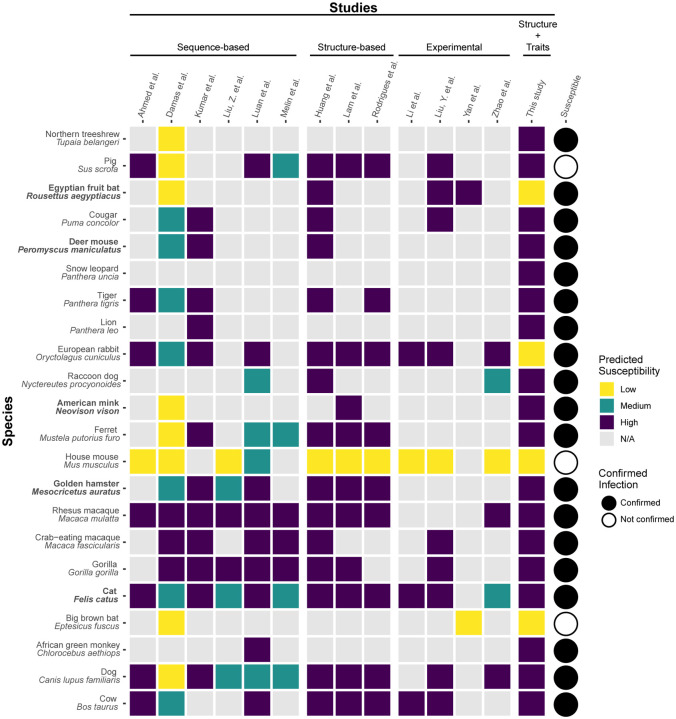
Figure 1.
A heatmap summarizing predicted susceptibility to SARS-CoV-2 for species with confirmed infection status from in vivo experimental studies or documented natural infections. Studies that make predictions about species susceptibility are shown in the x-axis, organized by method of prediction (those relying on ACE2 sequences, estimating binding strength using three dimensional structures, or laboratory experiments). Predictions about zoonotic capacity from this study are listed in the second to last column, with high and low categories determined by zoonotic capacity observed in Felis catus. Confirmed infections for species along the y-axis are summarized in (Gryseels et al., 2020) and are depicted as a series of filled or unfilled circles. Bolded species have been experimentally confirmed to transmit SARS-CoV-2 to naive conspecifics. Species predictions ranged from warmer colors (yellow: low susceptibility or zoonotic capacity for SARS-CoV-2) to cooler colors (purple: high susceptibility or zoonotic capacity). See supplementary file 1 for detailed methods about study categorization.
Sequence-based studies
Studies predicting host susceptibility based on amino acid sequence similarity between human (hACE2) and non-human ACE2 assume that a high degree of similarity is correlated with viral binding, especially at amino acid residues where hACE2 interacts with the SARS-CoV-2 spike glycoprotein. For some species, such as rhesus macaques (Deng et al., 2020), these qualitative predictions are borne out by in vivo studies (Figure 1) but predictions from these methods do not consistently match real-world outcomes. For example, sequence similarity predicted weak viral binding for minks and ferrets, which have all been confirmed as highly susceptible, with minks capable of onward transmission to conspecifics (Damas et al., 2020; Oude Munnink et al., 2020; Shi et al., 2020) (Figure 1). These mismatches to experimental or real-world outcomes may arise in part because protein three-dimensional structure, the main determinant of protein function, is robust to changes in amino acid sequence (Rodrigues et al., 2013; Sander and Schneider, 1991). As such, sequence alone does not capture the details of the ACE2 receptor interaction with the SARS-CoV-2 spike protein.
Structure-based studies
Modeling the three-dimensional structure of protein-protein complexes addresses some of the limitations of sequence-based approaches, and has proven useful to predict how different ACE2 orthologs bind to the SARS-CoV-2 viral spike protein receptor-binding domain (RBD) (S. D. Lam et al., 2020; Rodrigues et al., 2020). They can also be used to identify ACE2 amino acid residues essential for a productive interaction with the viral RBD, and thus improve predictive models of susceptibility through structure-based inference (Rodrigues et al., 2020). These studies leveraged known structures of the hACE2 receptor bound to the SARS-CoV-2 RBD and used powerful simulation methods to predict how variation across different ACE2 orthologs affects binding to the viral RBD. While these approaches successfully predicted strong binding for species that have been infected (e.g. domestic cat, tiger, dog, and ferret), the results are also not consistently supported by experiments. For instance, while guinea pig ACE2 scored favorably among susceptible species in one of the studies (Rodrigues et al., 2020), this ortholog was shown experimentally not to bind to the SARS-CoV-2 RBD (Li et al., 2020).
Although structural modeling has produced the most accurate results to date, all currently available approaches for predicting the host range of SARS-CoV-2 are fundamentally constrained by the availability and quality of ACE2 sequences. ACE2 is ubiquitous across chordates, likely because of its role in several highly conserved physiological pathways (Fournier et al., 2012). Because it is so highly conserved, the majority of mammal species (>6,000 species) are likely to have ACE2 receptors, but there are many fewer sequences available from which to make predictions using existing modeling methods (~300 species). The functional importance of the ACE2 receptor suggests that it has evolved in association with other intrinsic organismal traits that are more easily observed and for which data are more widely available. These suites of correlated organismal traits may provide a robust statistical proxy that can be leveraged to predict suitable hosts for SARS-CoV-2. Previous trait-based analyses applied statistical (machine) learning techniques to accurately distinguish the zoonotic capacity of various organisms (Han et al., 2020, 2015; Yang and Han, 2018), and predict likely hosts for particular groups of related viruses (Han et al., 2019, 2016), predictions which have subsequently been validated through independent laboratory and field investigations (e.g., (Goldstein et al., 2018; Yang et al., 2017)).
Here, we combine molecular structural modeling of viral binding with machine learning of species-level traits to generate predictions about species’ zoonotic capacity for SARS-CoV-2 virus across over 5000 mammal species, expanding our predictive capacity by an order of magnitude (Figure 2). Crucially, this combined approach enables predictions for species whose ACE2 sequences are not available by leveraging information available from viral binding dynamics and biological traits of potential hosts. In our workflow (Figure 2), we first carry out structural modeling to quantify the binding strength of SARS-CoV-2 RBD for vertebrate species using published ACE2 amino acid sequences (Sorokina et al., 2020). We then collate species traits and apply machine learning to predict zoonotic capacity for 5,400 mammal species, determined by a conservative threshold of susceptibility and onward transmission capacity of SARS-CoV-2 reported by in vivo studies. Because COVID-19 is, at this time, primarily a disease affecting humans, spillback infection of SARS-CoV-2 from humans to animals is the most likely mode by which new host species will become established. Among mammal species with the highest predicted zoonotic capacity for SARS-CoV-2, we identify a subset of species for which the threat of spillback infection appears greatest due to geographic overlaps and opportunities for contact with humans in areas of high SARS-CoV-2 prevalence globally.
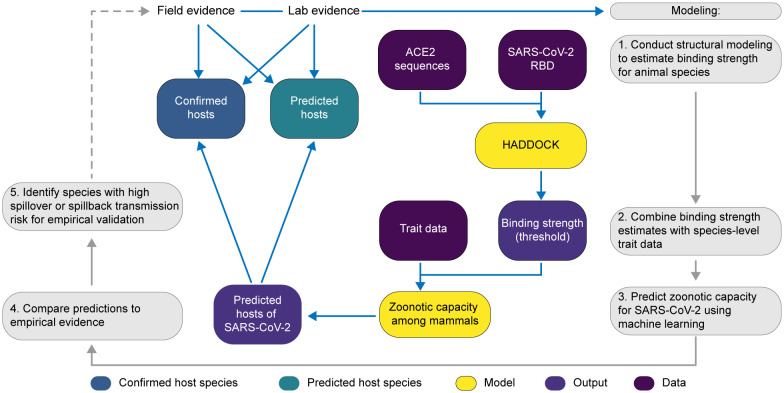
Figure 2.
A flowchart showing the progression of our workflow combining evidence from limited lab and field studies with additional data types to predict zoonotic capacity across mammals through multi-scale statistical modeling (gray boxes, steps 1–5). For all vertebrates with published ACE2 sequences, we modelled the interface of species’ ACE2 bound to the viral receptor binding domain using HADDOCK. We then combined the HADDOCK scores, which approximate binding strength, with species’ trait data and trained machine learning models for both mammals and vertebrates (yellow boxes). Mammal species predicted to have high zoonotic capacity were then compared to results of in vivo experiments and in silico studies that applied various computational approaches. We then identified a subset of species with particularly high risk of spillback and secondary spillover potential to prioritize additional lab validation and field surveillance (dashed line).
Methods
Protein sequence and alignment
We assembled a dataset of ACE2 NCBI GenBank accessions that are known human ACE2 orthologs or have high similarity to known orthologs as determined using BLASTx (Altschul et al., 1990). Using the R package rentrez and the accession numbers, we downloaded ACE2 protein sequences (Winter, 2017). We supplemented these sequences by manually downloading four additional sequences from the MEROPS database (Rawlings et al., 2018).
Structural Modeling of ACE2 orthologs bound to SARS-CoV-2 spike
The modeling of all 326 ACE2 orthologs bound to SARS-CoV-2 spike receptor binding domain was carried out as described previously (Rodrigues et al., 2020), with a few differences. In short, sequences of ACE2 orthologs were aligned using MAFFT (Katoh et al., 2002) and trimmed to the region resolved in the template crystal structure of hACE2 bound to the SARS-CoV-2 spike (PDB ID: 6m0j, (Lan et al., 2020). Ambiguous positions in each sequence, artifacts of the sequencing method, were replaced by Glycine to minimize assumptions about the nature of the amino acid side-chain but still allow for modeling. For each ortholog, we generated 10 homology models using MODELLER 9.24 (Sali and Blundell, 1993; Webb and Sali, 2016), with restricted optimization (fastest schedule) and refinement (very_fast schedule) settings, and selected a representative model based on the normalized DOPE score. These representative models were then manually inspected and 27 were removed from further analysis due to large insertions/deletions or to the presence of too many ambiguous amino acids at the interface with spike. Each validated model was submitted for refinement to the HADDOCK web server (van Zundert et al., 2016), which ran 50 independent short molecular dynamics simulations in explicit solvent to optimize the interface between the two proteins. For each one of the animal species in our study, we assigned an average and standard deviation of the scores of the 10 best refined models, ranked by their HADDOCK score -- a combination of van der Waals, electrostatics, and desolvation energies. A lower (more negative) HADDOCK score predicts stronger binding between the two proteins. We hereafter refer to predicted binding strength, or simply binding strength, to indicate HADDOCK score. The HADDOCK server is freely available, and we provide code to reproduce analyses or to aid in the application of this modeling approach to other similar problems (https://zenodo.org/record/4517509).
Trait data collection and cleaning
We gathered ecological and life history trait data from AnAge (de Magalhães and Costa, 2009), Amniote Life History Database (Myhrvold et al., 2015), and EltonTraits (Wilman et al., 2014), among other databases (supplementary file 2, Table 1; for details on data processing, see supplementary file 1 Methods). Using these data, we engineered additional traits that have shown importance in predicting host-pathogen associations in other contexts. For example, as a measure of habitat breadth (Dallas et al., 2017), we computed for each species the percentage of ecoregions it occupies. To assess the influence of sampling bias across species, we used the wosr R package (Baker, 2018) to count the number of studies returned in a search in Web of Science for each species’ Latin binomial and included this as a proxy for sampling bias in our model.
Modeling
Structure-based modeling of binding strength.
We began by modeling predicted binding strength for vertebrates, using boosted regression tree (BRT) models, an ensemble machine learning approach that accommodates non-random patterns of missing data, nonlinear relationships, and interacting effects among predictors. In a BRT model, a sequence of regression models are fit by recursive binary splits, with each additional regression modeling data that were poorly accounted for by the previous regression iterations in the tree (Elith et al., 2008). All BRT models were performed using the gbm package in R version 4.0.0 (Greenwell et al., 2020; R Core Team, 2020).
Quantifying a threshold for zoonotic capacity.
While ACE2 binding is necessary for viral entry into host cells, it is not sufficient for SARS-CoV-2 transmission. Multiple in vivo experiments suggest that not all species that are capable of binding SARS-CoV-2 are capable of transmitting active infection to other individuals (e.g., cattle, Bos taurus, (Ulrich et al., 2020); pigs, Sus scrofa, (Li et al., 2020)). Viral replication, and infectious viral shedding that enables onward transmission, are both required for a species to become a suitable bridge or reservoir species for SARS-CoV-2. In order to constrain our predictions to species with the potential to perpetuate onward transmission, we trained our models on a conservative threshold of binding strength (HADDOCK score = −129). Binding strength was binarized according to this threshold, above which it is more likely that both infection and onward transmission will occur following the results of multiple empirical studies (Table 1). This value is between the scores for two species: the domestic cat (Felis catus), which is currently the species with weakest predicted binding with confirmed conspecific transmission (Bosco-Lauth et al., 2020), and the pig (Sus scrofa), which shows the strongest estimated binding for which experimental inoculation failed to cause detectable infection (Shi et al., 2020). We note that there are species confirmed to be susceptible to SARS-CoV-2 whose predicted binding strength is weaker than cats, but conspecific transmission has not been confirmed in these species. While it is likely that intraspecific transmission will be reported for additional species, the binding strength selected for this analysis represents an appropriately conservative threshold based on currently available evidence. For additional modeling details, see supplementary file 1 Methods.
In addition, per-residue energy decomposition analysis of HADDOCK scores for 29 species indicated that all species with strong predicted binding had in common a salt bridge between SARS-CoV-2 K417 and a negatively charged amino acid at position 30 in the ACE2 sequence (Rodrigues et al., 2020). Given the apparent effect of amino acid 30 on overall binding strength, we constructed an additional feature to denote whether amino acid 30 is negatively charged (and therefore more likely to support strong binding) and included this feature as an additional trait in our models.
Trait-based modeling to predict zoonotic capacity
Prediction across multiple vertebrate classes is difficult due to extensive dissimilarities among traits describing different classes. For instance, traits that are commonly measured for reptiles are different than those of interest for birds or amphibians. Moreover, currently available ACE2 sequences are dominated by ray-finned fishes and mammals. Given that only mammals have so far been confirmed as both susceptible and capable of onward transmission of SARS-CoV-2, we created a separate set of models to make zoonotic capacity predictions for mammals only. For this mammal-only dataset, we gathered additional species-level traits from PanTHERIA (Jones et al., 2009) and added a series of binary fields for taxonomic order (based on (Wilson and Reeder, 2005); supplementary file 2, Table 2). We then applied boosted regression (BRT; gbm package, (Greenwell et al., 2020)) to impute missing trait data for mammal species (e.g., (Han et al., 2020); see supplementary file 1 Methods for details on imputation methods and results).
Many of the mammals for which we found the strongest evidence of zoonotic capacity are domesticated to some degree (pets, farmed or traded animals, lab models) (Oude Munnink et al., 2020; Schlottau et al., 2020; Shi et al., 2020). Relative to their ancestors or wild conspecifics, domesticated animals often have distinctive traits (Wilkins et al., 2014) that are likely to influence the number of zoonoses found in these species (Cleaveland et al., 2001). To account for trait variation due to domestication in certain species, we modeled mammals in two ways. First, we incorporated a variable indicating whether the source populations from which trait data were collected are wild or non-wild (e.g., farmed, pets, laboratory animals; non-wild status confirmed by the Mammal Diversity Database (Database, 2020)). Trait data collected from both wild and non-wild individuals were considered to represent non-wild species for the purposes of this model. In a second approach, we used only the wild species for model training and evaluation. For both approaches, pre-imputation trait values were used for all non-wild mammals during model training, evaluation, and prediction.
For boosted regression models, we applied grid search to select optimal hyperparameters, and repeated model fitting 50 times using bootstrapped training sets of 80% of labeled data. We measured performance by the area under the receiver operating characteristic curve (AUC) for predictions made on the test dataset (remaining 20%), corrected by comparing with null models created by target shuffling, which employed similar bootstrapping (50 times). Detailed methods can be found in supplementary file 1 Methods. We discuss herein the results of model predictions about zoonotic capacity made by applying this final model to all mammal species. We also report the mean and variation in predicted probabilities across all 50 bootstrapped models in supplementary file 4.
We identified mammal species with the top 10% of predicted probabilities of zoonotic capacity for SARS-CoV-2. We mapped the geographic ranges of these species using International Union for the Conservation of Nature (IUCN) polygons of species distributions (IUCN, 2020). We filtered this 90th percentile subset of mammal predictions to species that occur in human-associated habitats (e.g., urban areas, crop lands, pastures, heavily degraded forests) based on IUCN Red List assessments (IUCN 2020). We filtered a third time by masking the ranges of species that overlap with locations reporting cumulative human positive SARS-CoV-2 case data from the COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (Dong et al., 2020). While these cumulative case counts do not encompass the true extent of the pandemic due to uneven detection and reporting efforts across countries, they are currently the best available signal for the spread of SARS-CoV-2 at the global scale.
Additional methods and results of multiple uninformative model variations (e.g., a model in which binding strength is modeled as a continuous rather than a threshold measure, a model predicting the charge at amino acid 30) are also described in supplementary file 1 Methods and supplementary file 3 Table 3. Details about how predictions made by past studies were standardized into categories (low, medium, high; Figure 1) are also available in supplementary file 1 Methods.
Results
ACE2 host protein sequences and alignment
The ACE2 protein sequence alignment of the orthologs from 326 species spans eight classes and 87 orders (https://zenodo.org/record/4517509). The majority of sequences belonged to the classes Actinopterygii (22.1%), Aves (23.3%), and Mammalia (46.6%). Sequence length ranged from 344 amino acids to 872 with a median length of 805.
Structural modeling of viral binding strength
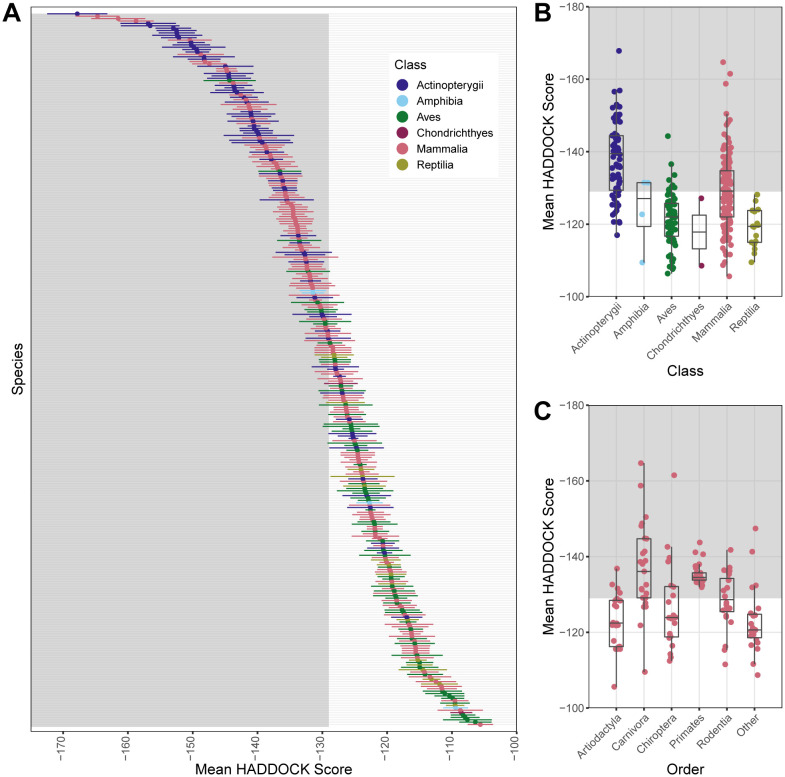
We predicted binding strength for 299 vertebrates, including 142 mammals. These binding strength scores represented six classes and 80 orders. Across these six vertebrate classes, the strongest predicted binding between ACE2 and SARS-CoV-2 (corresponding to the lowest mean HADDOCK scores), were in ray-finned fishes (Actinopterygii; mean = −137.945) and mammals (Mammalia; mean = −129.193) (Figure 3A). Each of these six classes included at least one species predicted to have stronger binding than Felis catus (Figure 3B). Overall, binding strength ranged from strongest binding observed for the cichlid Astatotilapia calliptera (−167.816) to weakest binding observed for alpaca (Vicugna pacos) (−105.615). Among well-represented mammalian orders (those containing at least 10 species with binding strength predictions), Primates and Carnivora showed predicted mean binding strengths that were stronger than domestic cats (Figure 3C).
Figure 3.
Plots showing results from modeling species’ ACE2 interaction with SARS-CoV-2 RBD using HADDOCK to predict binding strength (measured as arbitrary units, a.u.). HADDOCK scores that predict stronger binding are more negative. The mean and standard deviation of the HADDOCK score for vertebrate species (A) for which ACE2 orthologs are available. Binding strengths vary across vertebrate classes (B) and across the five most speciose mammalian orders (C). The “Other” category contains species across multiple orders for which ACE2 sequences were available, each with fewer than 10 representative species in the order. The shaded regions of all panels represent predicted binding that is as strong or stronger than (more negative values than) the domestic cat (Felis catus), which represents our conservative zoonotic capacity threshold based on currently available empirical evidence.
Trait-based machine learning models
Models created using the mammal-only dataset with trait imputation showed corrected test AUC of 0.72 for the zoonotic capacity model (for results of all models, see supplementary file 3). For mammal predictions, we applied the model trained on wild species given its higher accuracy (corrected test AUC = 0.72), compared to a model that included all available species and a variable indicating whether species trait data were collected from wild or non-wild individuals (corrected test AUC = 0.70). Citation count, as a proxy for study effort, had ~1% relative importance, suggesting that sampling bias across species had little influence on the model.
Species predictions of zoonotic capacity
The zoonotic capacity model identified 2,401 mammal species with prediction scores above 0.5, and 540 species within the 90th percentile probability (0.826 or higher), representing the subset of species assigned high confidence predictions of SARS-CoV-2 zoonotic capacity (similar to or greater than domestic cats). See supplementary file 4 for predictions on all 5,400 mammal species.
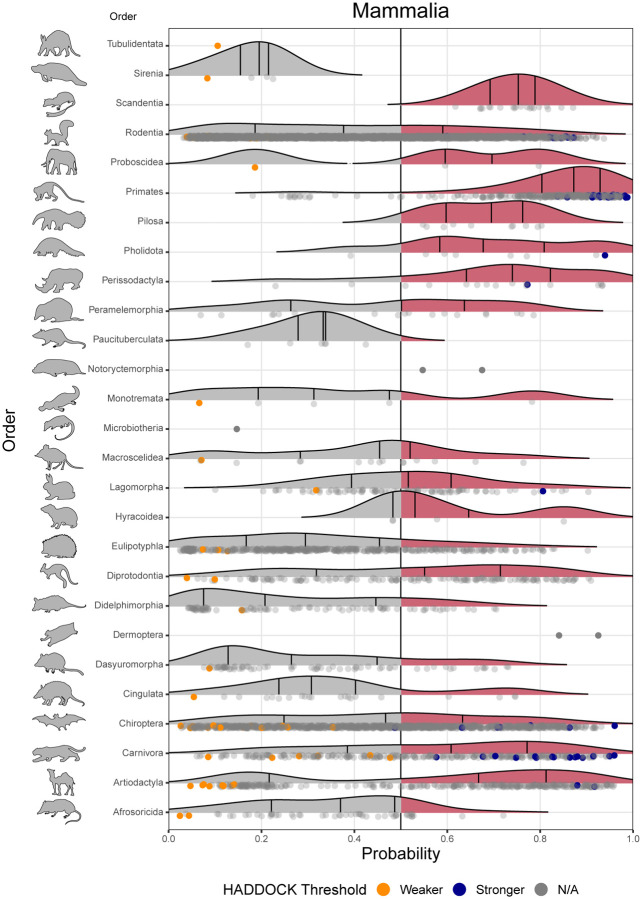
There were clear differences among mammalian orders in predicted zoonotic capacity. The top 10% of species with the highest predicted probabilities includes representatives from 13 orders. Most primates were predicted to have high zoonotic capacity and collectively showed stronger viral binding compared to other mammal groups (Figure 4). Additional orders with numerous species predicted to have high zoonotic capacity (at least 75% of species above 0.5) include Hyracoidea (hyraxes), Perissodactyla (odd-toed ungulates), Scandentia (treeshrews), Pilosa (sloths and anteaters), Pholidota (pangolins), and non-cetacean Artiodactyla (even-toed ungulates) (Figure 4).
Figure 4.
Ridgeline plots showing the distribution of predicted zoonotic capacity across mammals. Predicted probabilities for zoonotic capacity across the x-axis range from 0 (likely not susceptible) to 1 (zoonotic capacity predicted to be the same or greater than Felis catus), with the vertical line representing 0.5. The y-axis depicts all mammalian orders represented by our predictions. Density curves represent the distribution of the predictions, with those parts of the curve over 0.5 colored pink and lines representing distribution quartiles. The predicted values for each order are shown as points below the density curves. Points that were used to train the model are colored: orange represents species with weaker predicted binding, blue represents species with stronger predicted binding. Selected family-level distributions are shown in the two figure supplements for this figure.
Model predictions
Comparing species predictions across multiple computational approaches
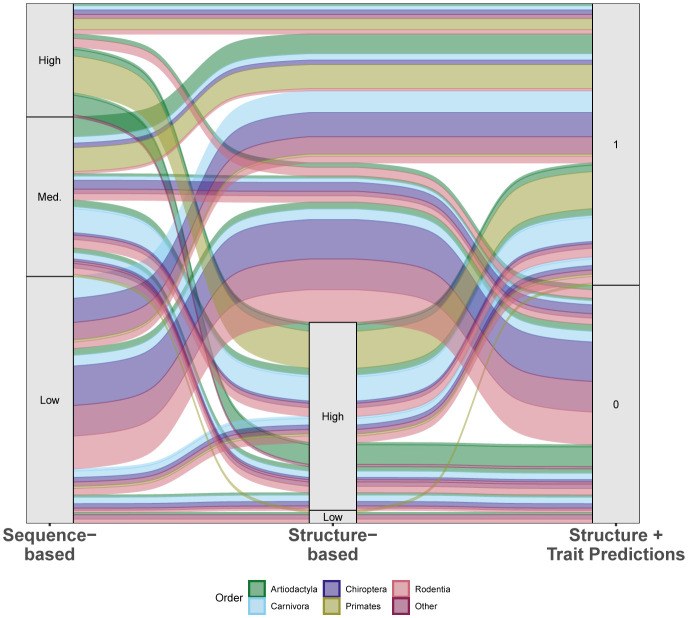
Our model combined species traits and viral binding strength to predict zoonotic capacity (susceptibility and onward transmission). We note that our threshold for zoonotic capacity was based on experimental studies confirming intraspecific transmission, and is therefore more conservative than thresholds adopted by other studies (e.g., based on binding strength, (Huang et al., 2020)). In addition, our modeling approach (machine learning) and prediction targets (zoonotic capacity) differed compared to existing computational approaches, which applied sequence-based or structure-based analyses that are limited to a small number of published ACE2 sequences. Despite these differences, comparing species predictions generated by multiple approaches can be useful for gauging consensus, and for comparing how predictions change from one method to another. Across multiple approaches there was general agreement in the predictions for primates and for a select group of artiodactyls and carnivores (Figure 5). Our model results also agree with some low susceptibility predictions made by several previous studies using sequence-based approaches (e.g., in certain bats and rodents). The structure-based models predicted a smaller proportion of species to have low susceptibility as compared to sequence-based studies.
Figure 5.
An alluvial plot comparing predictions of species susceptibility from multiple methods. Existing studies (listed in supplementary file 1 Methods) are categorized as either sequence-based or structure-based. Predictions from our zoonotic capacity model result from combining structure-based modeling of viral binding with organismal traits using machine learning to distinguish species with zoonotic capacity above (1) or below (0) a conservative threshold value set by domestic cats (Felis catus). Colors represent unique mammalian orders, and the width of colored bands representing the relative number of species with that combination of predictions across methods. See supplementary file 1 methods for details on how species across multiple studies were assigned to categories (high, medium, low).
Comparing species predictions to in vivo outcomes
Among the subset of species with ACE2 sequences (deposited in GenBank or MEROPS), our model predictions matched the results of most in vivo studies (Figure 1). For instance, model predictions were consistent with the results of numerous SARS-CoV-2 infection experiments on live animals. Experiments on deer mice (Peromyscus maniculatus; (Fagre et al., 2020; Griffin et al., 2020)) and raccoon dogs (Nyctereutes procyonoides; (Freuling et al., 2020)) confirmed SARS-CoV-2 infection and transmission to naive conspecifics. Our model also predicted a high probability of zoonotic capacity of American mink for SARS-CoV-2 (Neovison vison, probability=0.83, 90th percentile), in which farmed individuals present severe infection from human spillback, and demonstrate the capacity to transmit to conspecifics as well as to humans (Oreshkova et al., 2020; Oude Munnink et al., 2020). Our model also correctly predicted relatively low zoonotic capacity for big brown bats (Eptesicus fuscus; (Hall et al., 2020)) and house mice (wild type Mus musculus; (Bao et al., 2020)).
Some model predictions differed from the results of experimental studies. For instance, our model predicted a moderately high probability of zoonotic capacity for pigs (Sus scrofa, probability = 0.72, ~80th percentile). While some experiments have confirmed strong viral binding in this species (Li et al., 2020), others report no detectable infection or onward transmission of SARS-CoV-2 (Schlottau et al., 2020; Shi et al., 2020). Similarly for cattle (Bos taurus), our model also predicted a moderately high probability for zoonotic capacity (0.72, ~80th percentile), but in a live animal experiment, cattle were confirmed to be susceptible to infection but no transmission was observed to virus-naive conspecifics (Ulrich et al., 2020).
Mapping risk
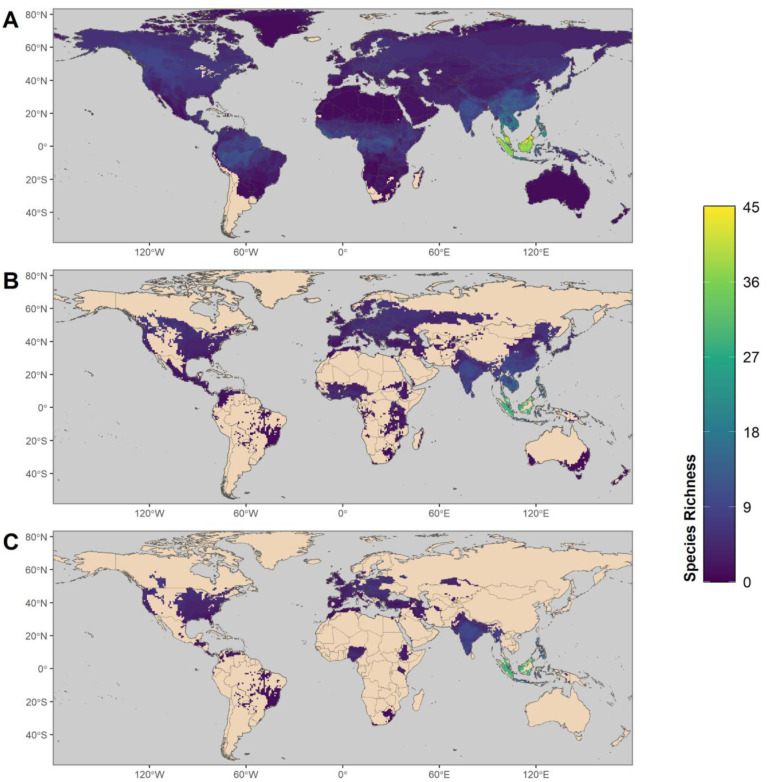
Most of the terrestrial world intersects the geographic range of at least one mammal species within the top 10% of predicted zoonotic capacity for SARS-CoV-2. The highest diversity of species within this top 10% occurs in the tropics (Figure 6A). Masking these species’ ranges to human-associated habitats showed that a total of 139 countries with at least one mammal species in the 90th percentile (Figure 6B). Restricting further to regions where there have been at least 100,000 cumulative human SARS-CoV-2 positive cases (as of 15 February 2021) highlighted 144 species across 71 countries (Figure 6C). These maps exclude the distributions of companion animals and zoo species, for which SARS-CoV-2 surveillance and veterinary records are not systematically available (McNamara et al., 2020). For a full list of model-predicted zoonotic capacity of species by country, see supplementary file 5.
Figure 6:
Maps showing the global distribution of species with predicted capacity to transmit SARS-CoV-2. (A) depicts global species richness of the top 10 percent of model-predicted zoonotic capacity. Ranges of this subset of species were filtered to those associated with human-dominated or human-altered habitats (B), and further filtered to show the subset of species that overlaps with areas of high human SARS-CoV-2 positive case counts (over 100,000 cumulative cases as of 15 February 2021) (C).
Discussion
We combined structure-based inference about viral binding with species-level trait data to make predictions about the capacity of animal species to become zoonotic hosts of SARS-CoV-2 (zoonotic capacity). Our definition of zoonotic capacity includes critical elements necessary for an animal host to serve as a zoonotic host, either as a new enzootic reservoir or as a bridge host capable of seeding secondary transmission to humans following an initial spillback event. First, species susceptibility to SARS-CoV-2 is a necessary condition, which we assumed to depend on the strength of binding between SARS-CoV-2 RBD and host ACE2. Second, the capacity for onward transmission, which we model as a threshold quantity based on available empirical evidence confirming SARS-CoV-2 transmission to naive conspecific hosts. To extend predictive capacity beyond the small number of species for which ACE2 sequences are currently available, we leveraged data on intrinsic biological traits of ~5400 mammal species. We assumed intrinsic traits to be under similarly broad selection pressures influencing major physiological pathways, such as those incorporating the ACE2 receptor across species. This combined modeling approach predicted zoonotic capacity with 72% accuracy, and identified numerous mammal species whose predicted zoonotic capacity meets or exceeds the viral susceptibility and transmissibility observed in experimental infections with SARS-CoV-2. In addition to wide agreement with in vivo study results (Table 1), model predictions corroborate multiple previous studies investigating species susceptibility to SARS-CoV-2 using the limited number of currently available ACE2 sequences (Figure 1).
Captive, farmed, or domesticated species.
Given that the type and frequency of contact with humans fundamentally underlies transmission risk, it is notable that our model predicted high zoonotic capacity for multiple captive species that have also been confirmed as susceptible to SARS-CoV-2 via experiments or natural infections. These include numerous carnivore species, such as large cats from multiple zoos, pet dogs and cats. Our model also predicted high SARS-CoV-2 zoonotic capacity for many farmed, domesticated, and live traded animal species. The water buffalo (Bubalus bubalis), widely bred for dairy production and farming, had the highest probability of zoonotic capacity among livestock (0.91). The 90th percentile of model predictions also included American mink (Neovison vison), red fox (Vulpes vulpes), sika deer (Cervus nippon), white-lipped peccary (Tayassu pecari), nilgai (Boselaphus tragocamelus), and raccoon dogs (Nyctereutes procyonoides), all of which are farmed, with the latter two considered invasive species in some areas (Milla et al., 2018; Pitra et al., 2010). In addition to the risks of secondary spillover to humans and the potential for large economic losses from culling infected animals (Kevany, 2020), the escape of farmed individuals into wild populations has implications for the spread and enzootic establishment of SARS-CoV-2 (DeLiberto and Shriner, 2020). These findings also have implications for informing vaccination strategies for people in regular contact with potential bridge species (e.g., veterinarians, abattoir-workers, farmers, etc).
Live traded or hunted wildlife species.
Model predictions also included many live-traded mammals. The majority of the legal live mammal trade consists of primates and carnivores (Can et al., 2019). Most live-traded primates come from the genus Macaca, with 20 out of 21 species in the genus predicted to have high zoonotic capacity, along with several live-traded carnivores, such as the Asiatic black bear (Ursus thibetanus), grey wolf (Canis lupus), and jaguar (Panthera onca). Two species of live-traded pangolins, the Philippine pangolin (Manis culionensis) and Sunda pangolin (M. javanica) were also predicted with high zoonotic capacity. Pangolins are notable because one of the betacoronaviruses with the highest sequence similarity to SARS-CoV-2 was isolated from Sunda pangolins (Andersen et al., 2020; T. T.-Y. Lam et al., 2020). Additional species in the top 10% of predictions that are commonly hunted include duiker (Cephalophus zebra, West Africa), warty pig (Sus celebes, Southeast Asia), and two species of deer (Odocoileus hemionus and O. virginianus) that are widespread across the Americas. The white-tailed deer (O. hemionus) was recently confirmed capable of transmitting SARS-CoV-2 to conspecifics via indirect contact (aerosolized virus particles) (Palmer et al., 2021).
Bats.
Similarly, bats are of special interest because of the high diversity of betacoronaviruses found in Rhinolophus spp. and other bat species (Anthony et al., 2017, 2013; Olival et al., 2020; Tsuda et al., 2012). Our model identified 35 bat species within the 90th percentile of zoonotic capacity for SARS-CoV-2. Within the genus Rhinolophus, our model identified the large rufous horseshoe bat (Rhinolophus rufus), a known natural host for bat betacoronaviruses (Tsuda et al., 2012) and a congener to three other horseshoe bats harboring betacoronaviruses with high nucleotide sequence similarity to SARS-CoV-2 (~92–96%) (Hul et al., 2021; H. Zhou et al., 2020; P. Zhou et al., 2020). For these three species, our model assigned a range of probabilities for SARS-CoV-2 zoonotic capacity (Rhinolophus affinis (0.58), R. malayanus (0.70), and R. shameli (0.71)). Our model identified additional congeners, Rhinolophus acuminatus (0.84) and R. macrotis (0.70), predicted to have relatively high probabilities. These predictions are in agreement with recent experiments demonstrating efficient viral binding of SARS-CoV-2 RBD for R. macrotis (Mou et al., 2020) and confirmation of SARS-CoV-2-neutralizing antibodies in field-caught R. acuminatus harboring a closely related betacoronavirus (Wacharapluesadee et al., 2021). Within the genus Pteropus (flying foxes), our model identified 17 species with high probabilities of zoonotic capacity for SARS-CoV-2. Some of these species are confirmed reservoirs of other zoonotic viruses in Southeast Asia (e.g., henipaviruses in P. lylei, P. vampyrus, P. conspicillatus, and P. alecto). While contact patterns between bats and humans may be somewhat less direct compared with captive or farmed species, annual outbreaks attributed to viral spillover transmission from bats illustrate a persistent epizootic risk to humans (Kessler et al., 2018; Plowright et al., 2015; Pulliam et al., 2012) and suggest that gaps in systematic surveillance of zoonotic viruses, including betacoronaviruses, are an urgent priority (e.g., (Peel et al., 2020)).
Rodents.
Our model identified 76 rodent species with high zoonotic capacity for SARS-CoV-2, some of which thrive in human-altered settings. Among these, our model predicted high probabilities for the deer mouse (Peromyscus maniculatus) and the white-footed mouse (P. leucopus). These are among the most well-studied mammals in North America, in part due to their status as zoonotic reservoirs for multiple zoonotic pathogens and parasites (Bordes et al., 2015; Machtinger and Williams, 2020; Ostfeld et al., 2006). Experimental infection, viral shedding, and sustained intraspecific transmission of SARS-CoV-2 were recently confirmed for P. maniculatus (Fagre et al., 2020; Griffin et al., 2020). Our model predicted low zoonotic capacity for Mus musculus (0.11), corresponding with recent in vivo experiments suggesting this species is not susceptible to infection by SARS-CoV-2 (Bao et al., 2020). Also in the top 10% were two rodent species considered to be human commensals whose geographic ranges are expanding due to human activities: Rattus argentiventer (0.84) and R. tiomanicus (0.79) (supplementary file 5) (Hamdan et al., 2017; Louys et al., 2020; Morand et al., 2015). Additional common rodent species with relatively high probabilities of zoonotic capacity include domesticated guinea pigs (Cavia porcellus), gerbils (Gerbillus gerbillus, Meriones tristrami), and several common mouse species (Apodemus peninsulae, A. flavicollis, and A. sylvaticus), all of which are known reservoirs for other zoonotic diseases. It is notable that many of these rodent species are regularly preyed upon by carnivore species, such as the red fox (Vulpes vulpes) or domestic cats (Felis catus) who themselves are likely to have high zoonotic capacity for SARS-CoV-2.
Species with large geographic ranges.
With sufficient opportunity for infectious contact, the risk of zoonotic spillback transmission increases with SARS-CoV-2 prevalence in human populations. Among species with high model-predicted zoonotic capacity, there were several relatively common species with very large geographic ranges or synanthropic tendencies that overlap with high prevalence global hotspots of COVID-19 (Figure 6, supplementary file 5). Notable species that are widely distributed across much of the northern hemisphere include the red fox (Vulpes vulpes, ~50 countries), the European polecat (Mustela putorius), the raccoon dog (Nyctereutes procyonoides), stoat (Mustela erminea) and wolf (Canis lupus). White-tailed deer (Odocoileus virginianus) are among the most geographically widespread species across Latin American countries with high SARS-CoV-2 prevalence. Globally, South and Southeast Asia had the highest diversity of mammal species with high predicted zoonotic capacity for SARS-CoV-2 (~90 species). Notable examples in the 90th percentile probability in this region include both rodents and bats. For example, Finlayson’s squirrel (Callosciurus finlaysonii) is native to Mainland Southeast Asia, but introductions via the pet trade in Europe have led to invasive populations in multiple countries (Bertolino and Lurz, 2013). Hunting has been documented for numerous bat species with geographic ranges across Southeast Asia (e.g., Cheiromeles torquatus, Cynopterus brachyotis, Rousettus amplexicaudatus, Macroglossus minimus) (Mildenstein et al., 2016; Ransaleleh et al., 2020), and there were multiple additional bat species in the 90th percentile probability from Asia and Africa where bats are subject to hunting and from which other betacoronaviruses have been identified (Anthony et al., 2017; Tampon et al., 2020). There were also several wide-ranging species whose contact with humans are limited to specialized settings. For instance, biologists and wildlife managers handle live individuals for research purposes, including grizzly bear (Ursus arctos), polar bear (Ursus maritimus), and wolf (Canis lupus), all of which are in the 89th percentile or above for predicted zoonotic capacity.
Other high priority mammal species.
Species that are in frequent contact with humans that showed more equivocal predictions warrant further investigation. For instance, while species such as horses (Equus caballus), goats (Capra hircus), and guinea pigs (Cavia porcellus) are not in the top 10% of predicted zoonotic capacity, due to the nature of their contact with humans they may experience greater risks of spillback infection, or pose a greater risk to humans for secondary spillover infection compared to many wild species. Conversely, while certain endangered or nearly extinct species are predicted to have relatively high zoonotic capacity, they may have fewer opportunities for human contact. For these species, populations that are under active conservation management may be at greater risk of spillback transmission. These species include the scimitar-horned oryx (Oryx dammah), addax (Addax nasomaculatus), and mountain gorillas (Gorilla beringei), in which spillback infection may occur through close-proximity eco-tourism activities (Weber et al., 2020). Indeed, spillback transmission of SARS-CoV-2 has already been confirmed in a closely related species, the Western lowland gorilla (Gorilla gorilla) in captivity (Gibbons, 2021). These species may benefit from focused risk mitigation efforts, such as those enacted recently to protect endangered black-footed ferrets (Mustela nigripes) from potential SARS-CoV-2 spillback (Aleccia, 2020).
All fifteen species of Tupaia treeshrews were predicted by our model to have medium to high probability (ranging from 0.62 to 0.87). One species, T. belangeri, has been explored as a potential lab model for several human infectious diseases including SARS-CoV-2 (Xu et al., 2020). Relative to other treeshrews, our model assigned only medium probability for SARS-CoV-2 zoonotic capacity in T. belangeri (0.67), which matches lab studies reporting asymptomatic infection and low viral shedding in this species (Zhao et al., 2020). In contrast, the common treeshrew (T. glis) was in the 94th percentile of zoonotic capacity (0.87 probability). These two species are sympatric in parts of their range, exist in close proximity to humans, and also overlap geographically with COVID-19 hotspots in Southeast Asia, suggesting the possibility of spillover transmission among congeners if spillback transmission occurs from humans to these species.
Strengthening predictive capacity for zoonoses.
While there was wide agreement between our model predictions and empirical studies, examining mismatches between experimental results and model-generated predictions may better focus research attention on characterizing what external conditions may be driving disconnects between predicted and observed zoonotic capacity. For instance, in pigs (Sus scrofa) multiple computational and experimental studies predicted susceptibility to SARS-CoV-2 (Figure 1), but this prediction has not been supported by results from whole animal inoculations, which so far have showed unproductive infection (Schlottau et al., 2020; Shi et al., 2020). Similarly, previous studies made contrasting predictions about SARS-CoV-2 susceptibility of American mink (Damas et al., 2020; S. D. Lam et al., 2020), Figure 1) whose very high zoonotic capacity was only confirmed ipso facto in multiple countries (Zhou and Shi, 2021).
Disconnects between real-world observations and in silico predictions of zoonotic capacity may arise because host susceptibility and transmission capacity are necessary but not sufficient for high zoonotic risk to be realized in natural settings. These processes depend strongly on the cellular environments in which cell entry and viral replication take place (e.g., the presence of suitable receptors and key proteases, (Letko et al., 2020)), and on host immunogenicity (Bean et al., 2013). These processes are therefore embedded in a broader ecological context impacting intra-host infection dynamics (latency, recrudescence, tolerance), and environmental drivers of host susceptibility and viral persistence that collectively determine where and when spillover may occur (Bean et al., 2013; Becker et al., 2019; Morris et al., 2020; Plowright et al., 2017). Insofar as data limitations (e.g., limited ACE2 sequences or species trait data) preclude perfect computational predictions of zoonotic capacity, laboratory experiments are also limited in assessing true zoonotic capacity. For SARS-CoV-2 and other host-pathogen systems, animals that are readily infected in the lab appear to be less susceptible in non-lab settings (ferrets in the lab vs. mixed results in ferrets as pets (OIE, 2021; Sawatzki et al., 2020; Schlottau et al., 2020); rabbits in the lab vs. rabbits as pets (Mykytyn et al., 2021; Ruiz-Arrondo et al., 2020)). Moreover, wildlife hosts that are confirmed to shed multiple zoonotic viruses in natural settings (e.g., bats, (Peel et al., 2019)) can be much less tractable for laboratory investigations (for instance, requiring high biosecurity containment and very limited sample sizes). While laboratory experiments are critical for understanding mechanisms of pathogenesis and disease, without field surveillance and population-level studies they are only partial reflections of zoonotic capacity in the natural world. These examples illustrate that there is no single methodology sufficient to understand and predict zoonotic transmission, for SARS-CoV-2 or any zoonotic pathogen, and further demonstrate the need for coordination among theoretical and statistical models, lab work, and field work to improve zoonotic predictive capacity (Restif et al., 2012). As new SARS-CoV-2 variants continue to emerge, our work demonstrates the utility of combining molecular structural modeling with machine learning for predicting future animal hosts, and the potential for similar multi-scale methods to bridge the many advances in molecular and structural modeling with ecological and biological data to extend predictive capacity for zoonotic pathogens whose host ranges remain uncharacterized due to persistent bottlenecks in field-collected data on wild hosts and their potentially zoonotic viruses. Integration of multiple methodologies, as done here, and more efficient iteration between computational predictions, laboratory experiments, and targeted animal surveillance will better link transmission mechanisms to the broader conditions enabling spillover, spillback, and secondary transmission in nature.
Supplementary Material
Acknowledgments
We are grateful for discussions with Drs. Alexandre Bonvin, Colin Parrish, Dennis Bente, Hyunwook Lee, Kathryn Hanley, Susan Hafenstein, and John Paul Schmidt about various components of this project. This work was supported by the NSF EEID program (DEB 1717282), DARPA PREEMPT program (D18AC00031), CREATE-NEO, a member of the NIH NIAID CREID program (1U01 AI151807-01), and the NVIDIA Corporation GPU grant program (BAH); by the NSF Polar program (OPP 1935870, 1947040) (AV); and by NIH NIGMS (R35GM122543) (JPGLMR).
Footnotes
Competing interests
The authors declare no competing interests.
References
- Ahmed R, Hasan R, Siddiki AMAMZ, Islam MS. 2021. Host range projection of SARS-CoV-2: South Asia perspective. Infect Genet Evol 87:104670. doi: 10.1016/j.meegid.2020.104670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleccia J. 2020. “The Biggest Nemesis”: Black-Footed Ferrets Get Experimental Coronavirus Vaccine. Kaiser Health News. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. 2020. The proximal origin of SARS-CoV-2. Nat Med 26:450–452. doi: 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony SJ, Johnson CK, Greig DJ, Kramer S, Che X, Wells H, Hicks AL, Joly DO, Wolfe ND, Daszak P, Karesh W, Lipkin WI, Morse SS, PREDICT Consortium, Mazet JAK, Goldstein T. 2017. Global patterns in coronavirus diversity. Virus Evol 3:vex012. doi: 10.1093/ve/vex012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony SJ, Ojeda-Flores R, Rico-Chavez O, Navarrete-Macias I, Zambrana-Torrelio CM, Rostal MK, Epstein JH, Tipps T, Liang E, Sanchez-Leon M, Sotomayor-Bonilla J, Aguirre AA, Avila-Flores R, Medellin RA, Goldstein T, Suzan G, Daszak P, Lipkin WI. 2013. Coronaviruses in bats from Mexico. J Gen Virol 94:1028–1038. doi: 10.1099/vir.0.049759-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. 2018. wosr: Clients to the “Web of Science” and “InCites” APIs.
- Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, Qu Y, Li F, Lv Q, Wang W, Xue J, Gong S, Liu M, Wang G, Wang S, Song Z, Zhao L, Liu P, Zhao L, Ye F, Wang H, Zhou W, Zhu N, Zhen W, Yu H, Zhang X, Guo L, Chen L, Wang C, Wang Y, Wang X, Xiao Y, Sun Q, Liu H, Zhu F, Ma C, Yan L, Yang M, Han J, Xu W, Tan W, Peng X, Jin Q, Wu G, Qin C. 2020. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583:830–833. doi: 10.1038/s41586-020-2312-y [DOI] [PubMed] [Google Scholar]
- Bartlett SL, Diel DG, Wang L, Zec S, Laverack M, Martins M, Caserta LC, Killian ML, Terio K, Olmstead C, Delaney MA, Stokol T, Ivančić M, Jenkins-Moore M, Ingerman K, Teegan T, McCann C, Thomas P, McAloose D, Sykes JM, Calle PP. 2021. SARS-COV-2 INFECTION AND LONGITUDINAL FECAL SCREENING IN MALAYAN TIGERS (PANTHERA TIGRIS JACKSONI), AMUR TIGERS (PANTHERA TIGRIS ALTAICA), AND AFRICAN LIONS (PANTHERA LEO KRUGERI) AT THE BRONX ZOO, NEW YORK, USA. J Zoo Wildl Med 51:733–744. doi: 10.1638/2020-0171 [DOI] [PubMed] [Google Scholar]
- Bean AGD, Baker ML, Stewart CR, Cowled C, Deffrasnes C, Wang L-F, Lowenthal JW. 2013. Studying immunity to zoonotic diseases in the natural host - keeping it real. Nat Rev Immunol 13:851–861. doi: 10.1038/nri3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker DJ, Washburne AD, Faust CL, Pulliam JRC, Mordecai EA, Lloyd-Smith JO, Plowright RK. 2019. Dynamic and integrative approaches to understanding pathogen spillover. Philos Trans R Soc Lond B Biol Sci 374:20190014. doi: 10.1098/rstb.2019.0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino S, Lurz PWW. 2013. Callosciurussquirrels: worldwide introductions, ecological impacts and recommendations to prevent the establishment of new invasive populations: Worldwide introductions of Callosciurussquirrels. Mamm Rev 43:22–33. doi: 10.1111/j.1365-2907.2011.00204.x [DOI] [Google Scholar]
- Bordes F, Blasdell K, Morand S. 2015. Transmission ecology of rodent-borne diseases: New frontiers. Integr Zool 10:424–435. doi: 10.1111/1749-4877.12149 [DOI] [PubMed] [Google Scholar]
- Bosco-Lauth AM, Hartwig AE, Porter SM, Gordy PW, Nehring M, Byas AD, VandeWoude S, Ragan IK, Maison RM, Bowen RA. 2020. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc Natl Acad Sci U S A 117:26382–26388. doi: 10.1073/pnas.2013102117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can ÖE, D’Cruze N, Macdonald DW. 2019. Dealing in deadly pathogens: Taking stock of the legal trade in live wildlife and potential risks to human health. Glob Ecol Conserv 17:e00515. doi: 10.1016/j.gecco.2018.e00515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C-F, Loh CB, Foo YK, Shen S, Fielding BC, Tan THP, Khan S, Wang Y, Lim SG, Hong W, Tan Y-J, Fu J. 2006. ACE2 orthologues in non-mammalian vertebrates (Danio, Gallus, Fugu, Tetraodon and Xenopus). Gene 377:46–55. doi: 10.1016/j.gene.2006.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S, Laurenson MK, Taylor LH. 2001. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philos Trans R Soc Lond B Biol Sci 356:991–999. doi: 10.1098/rstb.2001.0889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas T, Park AW, Drake JM. 2017. Predicting cryptic links in host-parasite networks. PLoS Comput Biol 13:e1005557. doi: 10.1371/journal.pcbi.1005557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, Hiller M, Koepfli K-P, Pfenning AR, Zhao H, Genereux DP, Swofford R, Pollard KS, Ryder OA, Nweeia MT, Lindblad-Toh K, Teeling EC, Karlsson EK, Lewin HA. 2020. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci U S A 117:22311–22322. doi: 10.1073/pnas.2010146117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Database MD. 2020. Mammal Diversity Database. doi: 10.5281/zenodo.4139818 [DOI] [Google Scholar]
- Davies NG, Barnard RC, Jarvis CI, Kucharski AJ, Munday J, Pearson CAB, Russell TW, Tully DC, Abbott S, Gimma A, Waites W, Wong KLM, van Zandvoort K, CMMID COVID-19 Working Group, Eggo RM, Funk S, Jit M, Atkins KE, John Edmunds W. 2020. Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. medRxiv 2020.12.24.20248822. doi: 10.1101/2020.12.24.20248822 [DOI] [Google Scholar]
- DeLiberto T, Shriner S. 2020. ProMED (No. 20201213.8015608). International Society for Infectious Diseases. [Google Scholar]
- de Magalhães JP, Costa J. 2009. A database of vertebrate longevity records and their relation to other life-history traits. J Evol Biol 22:1770–1774. doi: 10.1111/j.1420-9101.2009.01783.x [DOI] [PubMed] [Google Scholar]
- Deng W, Bao L, Gao H, Xiang Z, Qu Y, Song Z, Gong S, Liu J, Liu J, Yu P, Qi F, Xu Y, Li F, Xiao C, Lv Q, Xue J, Wei Q, Liu M, Wang G, Wang S, Yu H, Liu X, Zhao W, Han Y, Qin C. 2020. Rhesus macaques can be effectively infected with SARS-CoV-2 via ocular conjunctival route. bioRxiv. doi: 10.1101/2020.03.13.990036 [DOI] [Google Scholar]
- Dong E, Du H, Gardner L. 2020. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 20:533–534. doi: 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elith J, Leathwick JR, Hastie T. 2008. A working guide to boosted regression trees. J Anim Ecol 77:802–813. [DOI] [PubMed] [Google Scholar]
- Fagre A, Lewis J, Eckley M, Zhan S, Rocha SM, Sexton NR, Burke B, Geiss BJ, Peersen O, Kading R, Rovnak J, Ebel GD, Tjalkens RB, Aboellail T, Schountz T. 2020. SARS-CoV-2 infection, neuropathogenesis and transmission among deer mice: Implications for reverse zoonosis to New World rodents. bioRxiv. doi: 10.1101/2020.08.07.241810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier D, Luft FC, Bader M, Ganten D, Andrade-Navarro MA. 2012. Emergence and evolution of the renin-angiotensin-aldosterone system. J Mol Med 90:495–508. doi: 10.1007/s00109012-0894-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freuling CM, Breithaupt A, Müller T, Sehl J, Balkema-Buschmann A, Rissmann M, Klein A, Wylezich C, Höper D, Wernike K, Aebischer A, Hoffmann D, Friedrichs V, Dorhoi A, Groschup MH, Beer M, Mettenleiter TC. 2020. Susceptibility of Raccoon Dogs for Experimental SARS-CoV-2 Infection. Emerg Infect Dis 26:2982–2985. doi: 10.3201/eid2612.203733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage KL, Kosoy MY. 2005. Natural history of plague: perspectives from more than a century of research. Annu Rev Entomol 50:505–528. doi: 10.1146/annurev.ento.50.071803.130337 [DOI] [PubMed] [Google Scholar]
- Garry RF. 2021. Mutations arising in SARS-CoV-2 spike on sustained human-to-human transmission and human-to-animal passage. Virological. https://virological.org/t/mutations-arising-in-sars-cov-2-spike-on-sustained-human-to-human-transmission-and-human-to-animal-passage/578
- Gibbons A. 2021. Captive gorillas test positive for coronavirus. Science. doi: 10.1126/science.abg5458 [DOI] [Google Scholar]
- Goldstein T, Anthony SJ, Gbakima A, Bird BH, Bangura J, Tremeau-Bravard A, Belaganahalli MN, Wells HL, Dhanota JK, Liang E, Grodus M, Jangra RK, DeJesus VA, Lasso G, Smith BR, Jambai A, Kamara BO, Kamara S, Bangura W, Monagin C, Shapira S, Johnson CK, Saylors K, Rubin EM, Chandran K, Lipkin WI, Mazet JAK. 2018. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat Microbiol 3:1084–1089. doi: 10.1038/s41564-018-0227-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell B, Boehmke B, Cunningham J, Developers GBM. 2020. Generalized Boosted Regression Models. Comprehensive R Archive Network (CRAN). [Google Scholar]
- Griffin BD, Chan M, Tailor N, Mendoza EJ, Leung A, Warner BM, Duggan AT, Moffat E, He S, Garnett L, Tran KN, Banadyga L, Albietz A, Tierney K, Audet J, Bello A, Vendramelli R, Boese AS, Fernando L, Robbin Lindsay L, Jardine CM, Wood H, Poliquin G, Strong JE, Drebot M, Safronetz D, Embury-Hyatt C, Kobasa D. 2020. North American deer mice are susceptible to SARS-CoV-2. bioRxiv. doi: 10.1101/2020.07.25.221291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryseels S, De Bruyn L, Gyselings R, Calvignac-Spencer S, Leendertz FH, Leirs H. 2020. Risk of human-to-wildlife transmission of SARS-CoV-2. Mamm Rev 8:e00373–17. doi: 10.1111/mam.12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth S, Visher E, Boots M, Brook CE. 2019. Host phylogenetic distance drives trends in virus virulence and transmissibility across the animal-human interface. Philos Trans R Soc Lond B Biol Sci 374:20190296. doi: 10.1098/rstb.2019.0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JS, Knowles S, Nashold SW, Ip HS, Leon AE, Rocke T, Keller S, Carossino M, Balasuriya U, Hofmeister E. 2020. Experimental challenge of a North American bat species, big brown bat (Eptesicus fuscus), with SARS-CoV-2. Transbound Emerg Dis. doi: 10.1111/tbed.13949 [DOI] [PubMed] [Google Scholar]
- Hamdan NES, Ng YL, Lee WB, Tan CS, Khan FAA, Chong YL. 2017. Rodent Species Distribution and Hantavirus Seroprevalence in Residential and Forested areas of Sarawak, Malaysia. Trop Life Sci Res 28:151–159. doi: 10.21315/tlsr2017.28.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer SA, Pauvolid-Corrêa A, Zecca IB, Davila E, Auckland LD, Roundy CM, Tang W, Torchetti M, Killian ML, Jenkins-Moore M, Mozingo K, Akpalu Y, Ghai RR, Spengler JR, Behravesh CB, Fischer RSB, Hamer GL. 2020. Natural SARS-CoV-2 infections, including virus isolation, among serially tested cats and dogs in households with confirmed human COVID-19 cases in Texas, USA. bioRxiv. doi: 10.1101/2020.12.08.416339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BA, Majumdar S, Calmon FP, Glicksberg BS, Horesh R, Kumar A, Perer A, von Marschall EB, Wei D, Mojsilović A, Varshney KR. 2019. Confronting data sparsity to identify potential sources of Zika virus spillover infection among primates. Epidemics 27:59–65. doi: 10.1016/j.epidem.2019.01.005 [DOI] [PubMed] [Google Scholar]
- Han BA, O’Regan SM, Paul Schmidt J, Drake JM. 2020. Integrating data mining and transmission theory in the ecology of infectious diseases. Ecol Lett 23:1178–1188. doi: 10.1111/ele.13520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BA, Schmidt JP, Alexander LW, Bowden SE, Hayman DTS, Drake JM. 2016. Undiscovered Bat Hosts of Filoviruses. PLoS Negl Trop Dis 10:e0004815. doi: 10.1371/journal.pntd.0004815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BA, Schmidt JP, Bowden SE, Drake JM. 2015. Rodent reservoirs of future zoonotic diseases. Proc Natl Acad Sci U S A 112:7039–7044. doi: 10.1073/pnas.1501598112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhang C, Pearce R, Omenn GS, Zhang Y. 2020. Identifying the Zoonotic Origin of SARS-CoV-2 by Modeling the Binding Affinity between the Spike Receptor-Binding Domain and Host ACE2. J Proteome Res 19:4844–4856. doi: 10.1021/acs.jproteome.0c00717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hul V, Delaune D, Karlsson EA, Hassanin A, Tey PO, Baidaliuk A, Gámbaro F, Tu VT, Keatts L, Mazet J, Johnson C, Buchy P, Dussart P, Goldstein T, Simon-Lorière E, Duong V. 2021. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. bioRxiv. doi: 10.1101/2021.01.26.428212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN. 2020. The IUCN Red List of Threatened Species.
- Jones KE, Bielby J, Cardillo M, Fritz SA, O’Dell J, Orme CDL, Safi K, Sechrest W, Boakes EH, Carbone C, Connolly C, Cutts MJ, Foster JK, Grenyer R, Habib M, Plaster CA, Price SA, Rigby EA, Rist J, Teacher A, Bininda-Emonds ORP, Gittleman JL, Mace GM, Purvis A. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals: Ecological Archives E090–184. Ecology 90:2648–2648. doi: 10.1890/08-1494.1 [DOI] [Google Scholar]
- Katoh K, Misawa K, Kuma K-I, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JFY, Sharp PM, Shaw GM, Peeters M, Hahn BH. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523–526. doi: 10.1126/science.1126531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler MK, Becker DJ, Peel AJ, Justice NV, Lunn T, Crowley DE, Jones DN, Eby P, Sánchez CA, Plowright RK. 2018. Changing resource landscapes and spillover of henipaviruses. Ann N Y Acad Sci 1429:78–99. doi: 10.1111/nyas.13910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevany S. 2020. Danish Covid mink cull and future disease fears will kill fur trade, say farmers. The Guardian. [Google Scholar]
- Kumar A, Pandey SN, Pareek V, Narayan RK, Faiq MA, Kumari C. 2020. Predicting susceptibility for SARS-CoV-2 infection in domestic and wildlife animals using ACE2 protein sequence homology. Zoo Biol. doi: 10.1002/zoo.21576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SD, Bordin N, Waman VP, Scholes HM, Ashford P, Sen N, van Dorp L, Rauer C, Dawson NL, Pang CSM, Abbasian M, Sillitoe I, Edwards SJL, Fraternali F, Lees JG, Santini JM, Orengo CA. 2020. SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals. Sci Rep 10:16471. doi: 10.1038/s41598-020-71936-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TT-Y, Jia N, Zhang Y-W, Shum MH-H, Jiang J-F, Zhu H-C, Tong Y-G, Shi Y-X, Ni X-B, Liao Y-S, Li W-J, Jiang B-G, Wei W, Yuan T-T, Zheng K, Cui X-M, Li J, Pei G-Q, Qiang X, Cheung WY-M, Li L-F, Sun F-F, Qin S, Huang J-C, Leung GM, Holmes EC, Hu Y-L, Guan Y, Cao W-C. 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583:282–285. doi: 10.1038/s41586-020-2169-0 [DOI] [PubMed] [Google Scholar]
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. doi: 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Letko M, Marzi A, Munster V. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5:562–569. doi: 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, Zhu J, Zhang Q, Wu J, Liu L. 2020. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol 92:595–601. doi: 10.1002/jmv.25726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang H, Tang X, Fang S, Ma D, Du C, Wang Y, Pan H, Yao W, Zhang R, Zou X, Zheng J, Xu L, Farzan M, Zhong G. 2020. SARS-CoV-2 and Three Related Coronaviruses Utilize Multiple ACE2 Orthologs and Are Potently Blocked by an Improved ACE2-Ig. J Virol 94. doi: 10.1128/JVI.01283-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louisville Zoo. 2020. Louisville Zoo Female Snow Leopard Tests Positive for SARS-CoV-2. https://louisvillezoo.org/louisville-zoo-female-snow-leopard-tests-positive-for-sars-cov-2-media-release/
- Louys J, Herrera MB, Thomson VA, Wiewel AS, Donnellan SC, O’Connor S, Aplin K. 2020. Expanding population edge craniometrics and genetics provide insights into dispersal of commensal rats through Nusa Tenggara, Indonesia. Rec Aust Mus 72:287–302. doi: 10.3853/j.2201-4349.72.2020.1730 [DOI] [Google Scholar]
- Luan J, Jin X, Lu Y, Zhang L. 2020. SARS-CoV-2 spike protein favors ACE2 from Bovidae and Cricetidae. J Med Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machtinger ET, Williams SC. 2020. Practical Guide to Trapping Peromyscus leucopus (Rodentia: Cricetidae) and Peromyscus maniculatus for Vector and Vector-Borne Pathogen Surveillance and Ecology. J Insect Sci 20. doi: 10.1093/jisesa/ieaa028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathavarajah S, Stoddart AK, Gagnon GA, Dellaire G. 2020. Pandemic danger to the deep: the risk of marine mammals contracting SARS-CoV-2 from wastewater. doi: 10.1101/2020.08.13.249904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara T, Richt JA, Glickman L. 2020. A Critical Needs Assessment for Research in Companion Animals and Livestock Following the Pandemic of COVID-19 in Humans. Vector Borne Zoonotic Dis 20:393–405. doi: 10.1089/vbz.2020.2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin AD, Janiak MC, Marrone F 3rd, Arora PS, Higham JP. 2020. Comparative ACE2 variation and primate COVID-19 risk. Commun Biol 3:641. doi: 10.1038/s42003-020-01370-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildenstein T, Tanshi I, Racey PA. 2016. Exploitation of Bats for Bushmeat and Medicine In: Voigt CC, Kingston T, editors. Bats in the Anthropocene: Conservation of Bats in a Changing World. Cham: Springer International Publishing. pp. 325–375. doi: 10.1007/978-3319-25220-9_12 [DOI] [Google Scholar]
- Milla R, Bastida JM, Turcotte MM, Jones G, Violle C, Osborne CP, Chacón-Labella J, Sosinski ÊE Jr, Kattge J, Laughlin DC, Forey E, Minden V, Cornelissen JHC, Amiaud B, Kramer K, Boenisch G, He T, Pillar VD, Byun C. 2018. Phylogenetic patterns and phenotypic profiles of the species of plants and mammals farmed for food. Nat Ecol Evol 2:1808–1817. doi: 10.1038/s41559-018-0690-4 [DOI] [PubMed] [Google Scholar]
- Morand S, Bordes F, Chen H-W, Claude J, Cosson J-F, Galan M, Czirják GÁ, Greenwood AD, Latinne A, Michaux J, Ribas A. 2015. Global parasite and Rattus rodent invasions: The consequences for rodent-borne diseases. Integr Zool 10:409–423. doi: 10.1111/17494877.12143 [DOI] [PubMed] [Google Scholar]
- Morris DH, Yinda KC, Gamble A, Rossine FW, Huang Q, Bushmaker T, Fischer RJ, Matson MJ, van Doremalen N, Vikesland PJ, Marr LC, Munster VJ, Lloyd-Smith JO. 2020. The effect of temperature and humidity on the stability of SARS-CoV-2 and other enveloped viruses. bioRxiv. doi: 10.1101/2020.10.16.341883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H, Quinlan BD, Peng H, Guo Y, Peng S, Zhang L, Davis-Gardner ME, Gardner MR, Crynen G, Voo ZX, Bailey CC, Alpert MD, Rader C, Choe H, Farzan M. 2020. Mutations from bat ACE2 orthologs markedly enhance ACE2-Fc neutralization of SARS-CoV-2. bioRxiv. doi: 10.1101/2020.06.29.178459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B, Avanzato VA, Rosenke R, Hanley PW, Saturday G, Scott D, Fischer ER, de Wit E. 2020. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 585:268–272. doi: 10.1038/s41586-020-2324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhrvold NP, Baldridge E, Chan B, Sivam D, Freeman DL, Ernest SKM. 2015. An amniote life-history database to perform comparative analyses with birds, mammals, and reptiles: Ecological ArchivesE096–269. Ecology 96:3109–3000. doi: 10.1890/15-0846r.1 [DOI] [Google Scholar]
- Mykytyn AZ, Lamers MM, Okba NMA, Breugem TI, Schipper D, van den Doel PB, van Run P, van Amerongen G, de Waal L, Koopmans MPG, Stittelaar KJ, van den Brand JMA, Haagmans BL. 2021. Susceptibility of rabbits to SARS-CoV-2. Emerg Microbes Infect 10:1–7. doi: 10.1080/22221751.2020.1868951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ODA. 2020. Mink at affected Oregon farm negative for SARS-CoV-2, wildlife surveillance continues.
- OIE. 2021. Events in animals: OIE - World Organisation for Animal Health. https://www.oie.int/en/scientific-expertise/specific-information-and-recommendations/questions-and-answers-on-2019novel-coronavirus/events-in-animals/
- Olival KJ, Cryan PM, Amman BR, Baric RS, Blehert DS, Brook CE, Calisher CH, Castle KT, Coleman JTH, Daszak P, Epstein JH, Field H, Frick WF, Gilbert AT, Hayman DTS, Ip HS, Karesh WB, Johnson CK, Kading RC, Kingston T, Lorch JM, Mendenhall IH, Peel AJ, Phelps KL, Plowright RK, Reeder DM, Reichard JD, Sleeman JM, Streicker DG, Towner JS, Wang L-F. 2020. Possibility for reverse zoonotic transmission of SARS-CoV-2 to freeranging wildlife: A case study of bats. PLoS Pathog 16:e1008758. doi: 10.1371/journal.ppat.1008758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreshkova N, Molenaar RJ, Vreman S, Harders F, Oude Munnink BB, Hakze-van der Honing RW, Gerhards N, Tolsma P, Bouwstra R, Sikkema RS, Tacken MG, de Rooij MM, Weesendorp E, Engelsma MY, Bruschke CJ, Smit LA, Koopmans M, van der Poel WH, Stegeman A. 2020. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill 25. doi: 10.2807/1560-7917.ES.2020.25.23.2001005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostfeld RS, Canham CD, Oggenfuss K, Winchcombe RJ, Keesing F. 2006. Climate, deer, rodents, and acorns as determinants of variation in lyme-disease risk. PLoS Biol 4:e145. doi: 10.1371/journal.pbio.0040145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, van der Spek A, Tolsma P, Rietveld A, Brouwer M, Bouwmeester-Vincken N, Harders F, Honing RH der, Wegdam-Blans MCA, Bouwstra RJ, GeurtsvanKessel C, van der Eijk AA, Velkers FC, Smit LAM, Stegeman A, van der Poel WHM, Koopmans MPG. 2020. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. doi: 10.1126/science.abe5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer MV, Martins M, Falkenberg S, Buckley A, Caserta LC, Mitchell PK, Cassmann ED, Rollins A, Zylich NC, Renshaw RW, Guarino C, Wagner B, Lager K, Diel DG. 2021. Susceptibility of white-tailed deer (Odocoileus virginianus) to SARS-CoV-2. bioRxiv. doi: 10.1101/2021.01.13.426628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel AJ, Field HE, Aravena MR, Edson D, McCallum H, Plowright RK, Prada D. 2020. Coronaviruses and Australian bats: a review in the midst of a pandemic. Aust J Zool. doi: 10.1071/ZO20046 [DOI] [Google Scholar]
- Peel AJ, Wells K, Giles J, Boyd V, Burroughs A, Edson D, Crameri G, Baker ML, Field H, Wang L-F, McCallum H, Plowright RK, Clark N. 2019. Synchronous shedding of multiple bat paramyxoviruses coincides with peak periods of Hendra virus spillover. Emerg Microbes Infect 8:1314–1323. doi: 10.1080/22221751.2019.1661217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitra C, Schwarz S, Fickel J. 2010. Going west—invasion genetics of the alien raccoon dog Nyctereutes procynoides in Europe. Eur J Wildl Res 56:117–129. doi: 10.1007/s10344-009-0283-2 [DOI] [Google Scholar]
- Plowright RK, Eby P, Hudson PJ, Smith IL, Westcott D, Bryden WL, Middleton D, Reid PA, McFarlane RA, Martin G, Tabor GM, Skerratt LF, Anderson DL, Crameri G, Quammen D, Jordan D, Freeman P, Wang L-F, Epstein JH, Marsh GA, Kung NY, McCallum H. 2015. Ecological dynamics of emerging bat virus spillover. Proceedings of the Royal Society B 282:20142124. doi: 10.1098/rspb.2014.2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, Parrish CR, McCallum H, Hudson PJ, Ko AI, Graham AL, Lloyd-Smith JO. 2017. Pathways to zoonotic spillover. Nat Rev Microbiol. doi: 10.1038/nrmicro.2017.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam JRC, Epstein JH, Dushoff J, Rahman SA, Bunning M, Jamaluddin AA, Hyatt AD, Field HE, Dobson AP, Daszak P. 2012. Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonosis. J R Soc Interface 9:89–101. doi: 10.1098/rsif.2011.0223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, Connor T, Peacock T, L. RD, Volz E. 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563
- Ransaleleh TA, Nangoy MJ, Wahyuni I, Lomboan A, Koneri R, Saputro S, Pamungkas J, Latinne A. 2020. Identification of bats on traditional market in dumoga district, North Sulawesi. IOP Conf Ser: Earth Environ Sci 473:012067. doi: 10.1088/17551315/473/1/012067 [DOI] [Google Scholar]
- Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. 2018. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res 46:D624–D632. doi: 10.1093/nar/gkx1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: A language and environment for statistical computing. Vienna, Austria. [Google Scholar]
- Restif O, Hayman DTS, Pulliam JRC, Plowright RK, George DB, Luis AD, Cunningham AA, Bowen RA, Fooks AR, O’Shea TJ, Wood JLN, Webb CT. 2012. Model-guided fieldwork: practical guidelines for multidisciplinary research on wildlife ecological and epidemiological dynamics. Ecol Lett. doi: 10.1111/j.1461-0248.2012.01836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Oude Munnink BB, de Meulder D, van Amerongen G, van den Brand J, Okba NMA, Schipper D, van Run P, Leijten L, Sikkema R, Verschoor E, Verstrepen B, Bogers W, Langermans J, Drosten C, Fentener van Vlissingen M, Fouchier R, de Swart R, Koopmans M, Haagmans BL. 2020. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 368:1012–1015. doi: 10.1126/science.abb7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JPGLM, Barrera-Vilarmau S, M C Teixeira J, Sorokina M, Seckel E, Kastritis PL, Levitt M. 2020. Insights on cross-species transmission of SARS-CoV-2 from structural modeling. PLoS Comput Biol 16:e1008449. doi: 10.1371/journal.pcbi.1008449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JPGLM, Melquiond ASJ, Karaca E, Trellet M, van Dijk M, van Zundert GCP, Schmitz C, de Vries SJ, Bordogna A, Bonati L, Kastritis PL, Bonvin AMJJ. 2013. Defining the limits of homology modeling in information-driven protein docking. Proteins 81:2119–2128. doi: 10.1002/prot.24382 [DOI] [PubMed] [Google Scholar]
- Ruiz-Arrondo I, Portillo A, Palomar AM, Santibanez S, Santibanez P, Cervera C, Oteo JA. 2020. Detection of SARS-CoV-2 in pets living with COVID-19 owners diagnosed during the COVID-19 lockdown in Spain: A case of an asymptomatic cat with SARS-CoV-2 in Europe. bioRxiv. doi: 10.1101/2020.05.14.20101444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. doi: 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Sander C, Schneider R. 1991. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins 9:56–68. doi: 10.1002/prot.340090107 [DOI] [PubMed] [Google Scholar]
- San Diego Zoo. 2021. Gorilla Troop at the San Diego Zoo Safari Park Test Positive for COVID-19. https://zoo.sandiegozoo.org/pressroom/news-releases/gorilla-troop-san-diego-zoo-safari-park-test-positive-covid-19
- Sawatzki K, Hill N, Puryear W, Foss A, Stone J, Runstadler J. 2020. Ferrets not infected by SARS-CoV-2 in a high-exposure domestic setting. bioRxiv. doi: 10.1101/2020.08.21.254995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, Höper D, Mettenleiter TC, Balkema-Buschmann A, Harder T, Grund C, Hoffmann D, Breithaupt A, Beer M. 2020. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe 1:e218–e225. doi: 10.1016/S2666-5247(20)30089-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, Chen H, Bu Z. 2020. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 368:1016–1020. doi: 10.1126/science.abb7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner S, Ellis J, Root JJ, Roug A, Stopak S, Wiscomb G, Zierenberg J, Ip H, Torchetti M, DeLiberto T. 2021. SARS-CoV-2 Exposure in Escaped Mink, Utah, USA. Emerging Infectious Disease journal 27. doi: 10.3201/eid2703.204444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia SF, Yan L-M, Chin AWH, Fung K, Choy K-T, Wong AYL, Kaewpreedee P, Perera RAPM, Poon LLM, Nicholls JM, Peiris M, Yen H-L. 2020. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583:834–838. doi: 10.1038/s41586-020-2342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit THC, Brackman CJ, Ip SM, Tam KWS, Law PYT, To EMW, Yu VYT, Sims LD, Tsang DNC, Chu DKW, Perera RAPM, Poon LLM, Peiris M. 2020. Infection of dogs with SARS-CoV-2. Nature 586:776–778. doi: 10.1038/s41586-020-2334-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokina M, M C Teixeira J, Barrera-Vilarmau S, Paschke R, Papasotiriou I, Rodrigues JPGLM, Kastritis PL. 2020. Structural models of human ACE2 variants with SARS-CoV-2 Spike protein for structure-based drug design. Sci Data 7:309. doi: 10.1038/s41597-020-00652-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tampon NVT, Rabaya YMC, Malbog KMA, Burgos SC, Libre K Jr, Valila ASD, Achondo MJMM, Onggo LS, Murao LAE. 2020. First molecular evidence for bat betacoronaviruses in Mindanao. Philipp J Sci 149:91–94. [Google Scholar]
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437:889. doi: 10.1038/nature04230 [DOI] [PubMed] [Google Scholar]
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, von Gottberg A, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao M, Korsman S, Davies M-A, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LCJ, Kosakovsky Pond SL, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C, de Oliveira T. 2020. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020.12.21.20248640. doi: 10.1101/2020.12.21.20248640 [DOI] [Google Scholar]
- Tsuda S, Watanabe S, Masangkay JS, Mizutani T, Alviola P, Ueda N, Iha K, Taniguchi S, Fujii H, Kato K, Horimoto T, Kyuwa S, Yoshikawa Y, Akashi H. 2012. Genomic and serological detection of bat coronavirus from bats in the Philippines. Arch Virol 157:2349–2355. doi: 10.1007/s00705-012-1410-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich L, Wernike K, Hoffmann D, Mettenleiter TC, Beer M. 2020. Experimental Infection of Cattle with SARS-CoV-2. Emerg Infect Dis 26:2979–2981. doi: 10.3201/eid2612.203799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA. 2020. Cases of SARS-CoV-2 in animals in the United States. https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/sa_one_health/sars-cov-2-animals-us
- Van Egeren D, Novokhodko A, Stoddard M, Tran U, Zetter B, Rogers M, Pentelute BL, Carlson JM, Hixon M, Joseph-McCarthy D, Chakravarty A. 2020. Risk of evolutionary escape from neutralizing antibodies targeting SARS-CoV-2 spike protein. bioRxiv. doi: 10.1101/2020.11.17.20233726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zundert GCP, Rodrigues JPGLM, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond ASJ, van Dijk M, de Vries SJ, Bonvin AMJJ. 2016. The HADDOCK2.2 Web Server: UserFriendly Integrative Modeling of Biomolecular Complexes. J Mol Biol 428:720–725. doi: 10.1016/j.jmb.2015.09.014 [DOI] [PubMed] [Google Scholar]
- Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ, Dabrera G, O’Toole Á, Amato R, Ragonnet-Cronin M, Harrison I, Jackson B, Ariani CV, Boyd O, Loman NJ, McCrone JT, Gonçalves S, Jorgensen D, Myers R, Hill V, Jackson DK, Gaythorpe K, Groves N, Sillitoe J, Kwiatkowski DP, The COVID-19 Genomics UK (COGUK) consortium, Flaxman, Ratmann, Bhatt S, Hopkins S, Gandy A, Rambaut A, Ferguson NM. 2021. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv 2020.12.30.20249034. doi: 10.1101/2020.12.30.20249034 [DOI] [Google Scholar]
- Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, Kaewpom T, Chia WN, Ampoot W, Lim BL, Worachotsueptrakun K, Chen VC-W, Sirichan N, Ruchisrisarod C, Rodpan A, Noradechanon K, Phaichana T, Jantarat N, Thongnumchaima B, Tu C, Crameri G, Stokes MM, Hemachudha T, Wang L-F. 2021. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat Commun 12:972. doi: 10.1038/s41467-021-21240-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Mitchell PK, Calle PP, Bartlett SL, McAloose D, Killian ML, Yuan F, Fang Y, Goodman LB, Fredrickson R, Elvinger F, Terio K, Franzen K, Stuber T, Diel DG, Torchetti MK. 2020. Complete Genome Sequence of SARS-CoV-2 in a Tiger from a U.S. Zoological Collection. Microbiol Resour Announc 9. doi: 10.1128/MRA.00468-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, Sali A. 2016. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics 54:5.6.1–5.6.37. doi: 10.1002/cpbi.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Kalema-Zikusoka G, Stevens NJ. 2020. Lack of Rule-Adherence During Mountain Gorilla Tourism Encounters in Bwindi Impenetrable National Park, Uganda, Places Gorillas at Risk From Human Disease. Front Public Health 8:1. doi: 10.3389/fpubh.2020.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2021. WHO coronavirus disease (COVID-19) dashboard.
- WHO. 2020. SARS-CoV-2 mink-associated variant strain – Denmark. World Health Organization. [Google Scholar]
- Wilkins AS, Wrangham RW, Fitch WT. 2014. The “domestication syndrome” in mammals: a unified explanation based on neural crest cell behavior and genetics. Genetics 197:795–808. doi: 10.1534/genetics.114.165423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilman H, Belmaker J, Simpson J, de la Rosa C, Rivadeneira MM, Jetz W. 2014. EltonTraits 1.0: Species-level foraging attributes of the world’s birds and mammals. Ecology 95:2027–2027. doi: 10.1890/13-1917.1 [DOI] [Google Scholar]
- Wilson DE, Reeder DM. 2005. Mammal Species of the World: A Taxonomic and Geographic Reference. Baltimore, MD: JHU Press. [Google Scholar]
- Winter D. 2017. rentrez: An R package for the NCBI eUtils API. R J 9:520. doi: 10.32614/rj-2017-058 [DOI] [Google Scholar]
- Woolsey C, Borisevich V, Prasad AN, Agans KN, Deer DJ, Dobias NS, Heymann JC, Foster SL, Levine CB, Medina L, Melody K, Geisbert JB, Fenton KA, Geisbert TW, Cross RW. 2020. Establishment of an African green monkey model for COVID-19. bioRxiv 2020.05.17.100289. doi: 10.1101/2020.05.17.100289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yu D-D, Ma Y-H, Yao Y-L, Luo R-H, Feng X-L, Cai H-R, Han J-B, Wang X-H, Li M-H, Ke C-W, Zheng Y-T, Yao Y-G. 2020. COVID-19-like symptoms observed in Chinese tree shrews infected with SARS-CoV-2. Zool Res 41:517–526. doi: 10.24272/j.issn.20958137.2020.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LH, Han BA. 2018. Data-driven predictions and novel hypotheses about zoonotic tick vectors from the genus Ixodes. BMC Ecol 18:7. doi: 10.1186/s12898-018-0163-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-L, Zhang Y-Z, Jiang R-D, Guo H, Zhang W, Li B, Wang N, Wang L, Waruhiu C, Zhou J-H, Li S-Y, Daszak P, Wang L-F, Shi Z-L. 2017. Genetically Diverse Filoviruses in Rousettus and Eonycteris spp. Bats, China, 2009 and 2015. Emerg Infect Dis 23:482–486. doi: 10.3201/eid2303.161119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang H, Huang K, Yang Y, Hui X, Gao J, He X, Li C, Gong W, Zhang Y, Peng C, Gao X, Chen H, Zou Z, Shi Z, Jin M. 2020. SARS-CoV-2 neutralizing serum antibodies in cats: a serological investigation. doi: 10.1101/2020.04.01.021196 [DOI] [Google Scholar]
- Zhao Y, Wang J, Kuang D, Xu J, Yang M, Ma C, Zhao S, Li J, Long H, Ding K, Gao J, Liu J, Wang H, Li H, Yang Y, Yu W, Yang J, Zheng Y, Wu D, Lu S, Liu H, Peng X. 2020. Susceptibility of tree shrew to SARS-CoV-2 infection. Sci Rep 10:16007. doi: 10.1038/s41598-020-72563-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, Wang P, Liu D, Yang J, Holmes EC, Hughes AC, Bi Y, Shi W. 2020. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr Biol 30:2196–2203.e3. doi: 10.1016/j.cub.2020.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Shi Z-L. 2021. SARS-CoV-2 spillover events. Science 371:120–122. doi: 10.1126/science.abf6097 [DOI] [PubMed] [Google Scholar]
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.