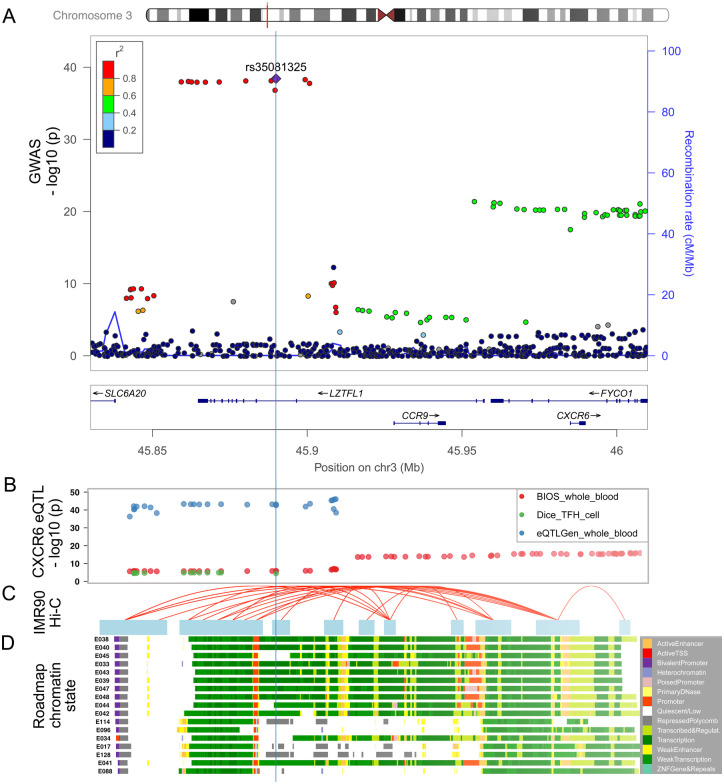
Fig. 3.
Functional genomic annotation on 3p21.31 locus with signals from GWASHGI.
(a) LocusZoom view of the association signals of SNPs at the 3p21.31 locus of GWASHGI. The x-axis is the chromosome position in million base pairs (Mb) on GRCh37 reference genome and y-axis represents the −log10 (p-value) from GWASHGI dataset. The color indicates the strength of linkage disequilibrium from the lead SNP rs35081325. The genes within the region are annotated in the lower panel. A vertical blue line labels the position of the lead SNP rs35081325 to denote the relationship of GWAS variants to other datasets: expression quantitative trait (eQTL) (Fig. 3b), chromatin interaction (Fig. 3c), and imputed Roadmap functional elements (Fig. 3d). (b) The significant eQTLs associated with CXCR6 expression in this region. The cis- eQTL datasets include two whole blood datasets [Biobank-based Integrative Omics Studies (BIOS) QTL and eQTLGen] and one T follicular helper cell dataset (DICE). The y axis represents the −log10 (p-value) from the eQTL studies. (c) The significant Hi-C interactions in normal lung fibroblast cell line (IMR90). Blue blocks denote the target and bait regions, and red arcs indicate the interactions between functional elements. (d) The region annotated with the chromatin-state segmentation track (ChromHMM) from the Roadmap Epigenomics data for T-cell and lung tissue. The Roadmap Epigenomics cell line IDs are shown on the left side: E017 (IMR90 fetal lung fibroblasts Cell Line), E033 (Primary T Cells from cord blood), E034 (Primary T Cells from blood), E038 (Primary T help naïve cells from peripheral blood), E039 (Primary T helper naïve cells from peripheral blood), E040 (Primary T helper memory cells from peripheral blood 1), E041 (Primary T helper cells PMA-Ionomycin stimulated), E042 (Primary T helper 17 cells PMA-Ionomycin stimulated), E043 (Primary T helper cells from peripheral blood), E044 (Primary T regulatory cells from peripheral blood), E045 (Primary T cells effector/memory enriched from peripheral blood), E047 (Primary T CD8 naïve cells from peripheral blood), E048 (Primary T CD8 memory cells from peripheral blood), E088 (Fetal lung), E096 (Lung), E114 (A549 EtOH 0.02pct Lung Carcinoma Cell Line), and E128 (NHLF Human Lung Fibroblast Primary Cells). The colors denote chromatin states imputed by ChromHMM, with the color key in the gray box (Methods).