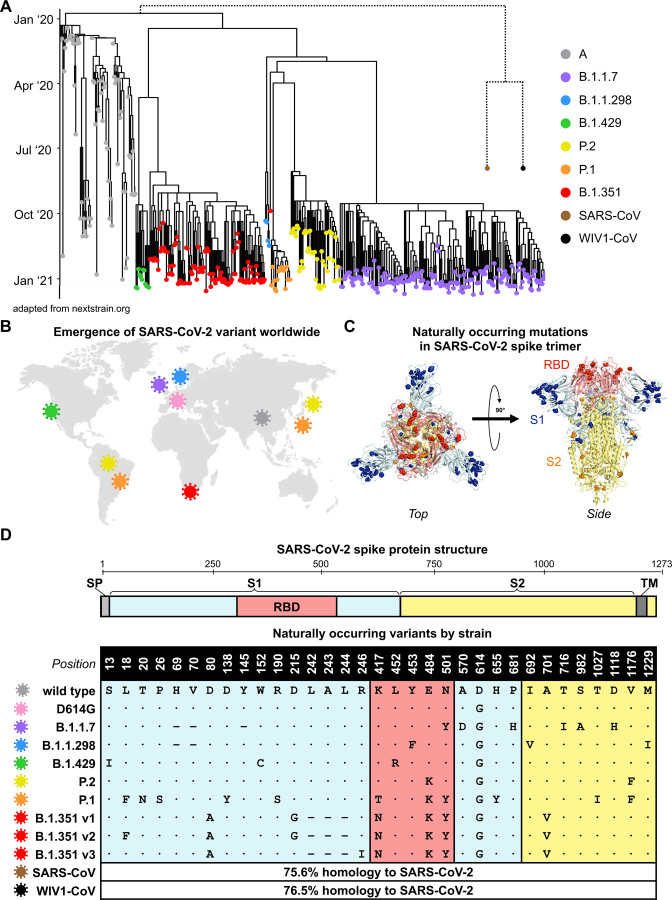
Figure 1. Emergence and spread of SARS-CoV-2 variants of concern around the world.
(A) Phylogenetic tree of SARS-CoV-2 variants (adapted from nextstrain.org) with sampling dates is illustrated with a focus on the following lineages: A (grey), B.1.1.7 (purple), B.1.1.298 (blue), B.1.429 (green), P.2 (yellow), P.1 (orange), and B.1.351 (red). Dotted lines to SARS-CoV (brown) and bat-derived WIV1-CoV (black) are not to scale but indicate a distant phylogenetic relationship to SARS-CoV-2.
(B) World map depicting the locations where the variants of these lineages were first described: original wild-type virus from A lineage (grey) in Wuhan, China; D614G variant (pink) in Europe that became dominant circulating strain; B.1.1.7 lineage (purple) in the United Kingdom; B.1.1.298 (blue) in Denmark; B.1.429 (green) in California, United States; P.2 (yellow) in Brazil and Japan; P.1 (orange) in Brazil and Japan; and B.1.351 (red) in South Africa.
(C) Crystal structure of pre-fusion stabilized SARS-Cov-2 spike trimer (PDB ID 7JJI) is shown with top (left panel) and side (right panel) views. Sites where naturally occurring mutations occur are indicated with residue atoms highlighted as colored spheres. Spike regions and associated mutations are colored as follows: receptor binding domain (RBD) in red, S1 (excluding RBD) in blue, and S2 in yellow.
(D) Schematic of SARS-CoV-2 spike protein structure and the mutation landscape of variants used in this study are illustrated. The mutations present in each variant tested represent the consensus sequence for that lineage and represent actual circulating strains: A (wild type), B.1.1.7, B.1.1.298, B.1.1.429, P.2, and P.1 lineages. In the case of the B.1.351 lineage, the three most abundant variants (v1, v2, and v3) deposited in GISAID were assessed. For SARS-CoV and WIV1-CoV, the percent homology is indicated. The following abbreviations are used: SP, signal peptide; TM, transmembrane domain; RBD, receptor binding domain. In the mutation map, a dot (∙) indicates the same amino acid in that position as wild type and a dash (–) indicates a deletion.