Figure 4.
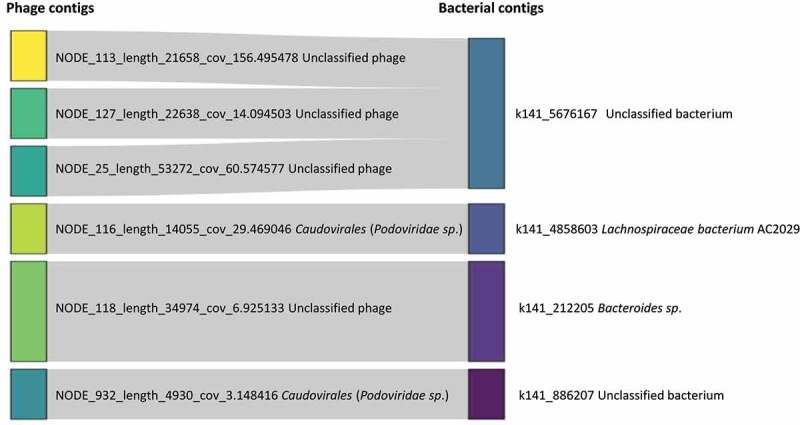
Sankey plot of putative phage contigs and their putative bacterial hosts based on CRISPR spacer sequences in VC_1832_0 and VC_1982_0, which were two of ten differentially abundant VCs between IBS and controls. To produce this plot, phage contigs in the 10 VCs were restricted to those that had a match based on CRISPR spacer sequences to a bacterial contig, resulting in 6 contigs, 4 from viral cluster, VC_1832_0 and 2 from VC_1982_0 (phage contigs beginning with NODE_116 and NODE_932). VC_1832_0 had significantly decreased abundance, and VC_1982_0 had significantly increased abundance in IBS compared with control samples, as reported by DeSeq2. For phage contigs, the putative species is indicated in brackets beside the phage family name
