Figure.
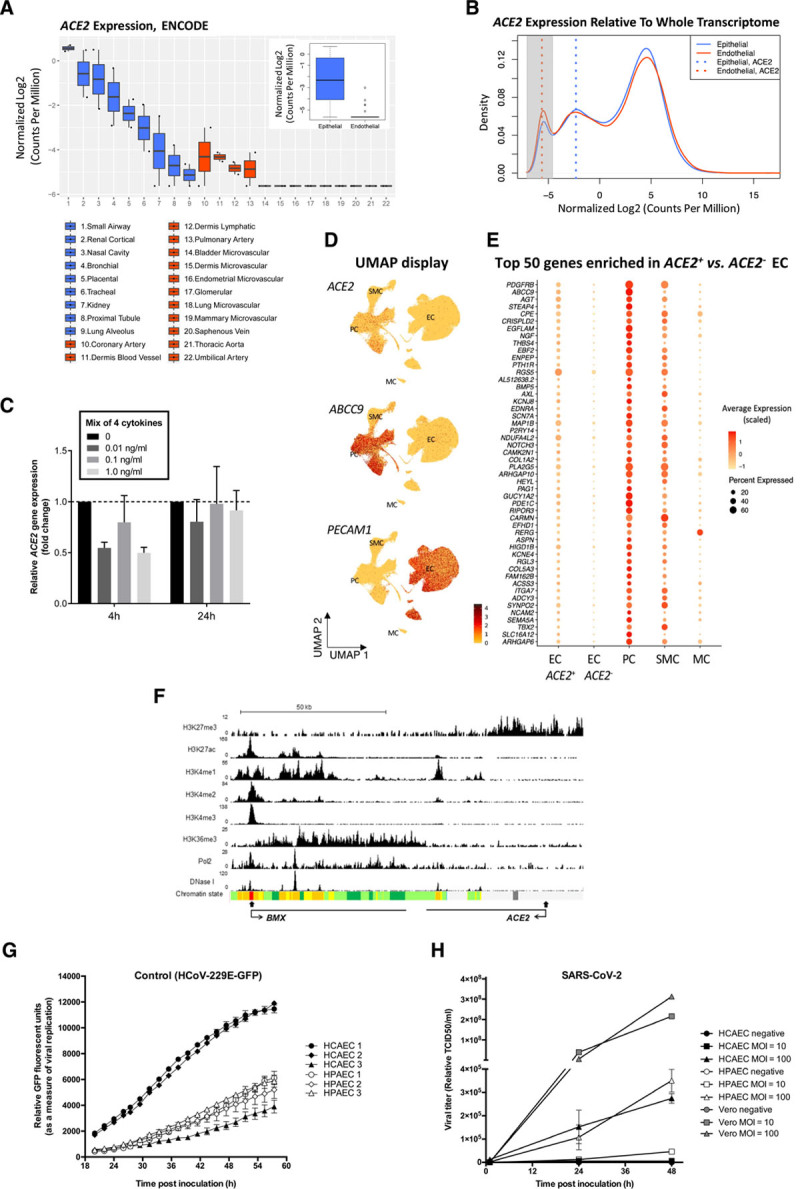
Analysis of ACE2 expression in human ECs and of coronavirus replication in primary human ECs. A and B, Comparison of ACE2 expression in human primary epithelial cells and ECs using total RNA sequencing data from the ENCODE database shows low or absent expression in ECs. A, The difference of ACE2 expression in epithelial cells and ECs is shown in boxplots with individual, as well as grouped, samples (inner boxplot). Each dot represents a single sample (n=2 per cell type). B, Transcriptome profiles of epithelial cells and ECs are shown in a density plot, using the median of all samples per group (n=19 360 genes). ACE2 expression in each group is marked with a dotted line: ACE2 expression in ECs (red) overlaps with the peak for nonexpressed transcripts (highlighted in grey), while ACE2 expression in epithelial cells (blue) is to the right, indicating detectable expression. Median ACE2 expression in endothelial cells equals -5.6 in log2 CPM, which corresponds with 0 raw read counts, signifying undetectable ACE2 expression in the majority of ECs. Expression values in all plots are represented as log2-transformed CPM, normalized by trimmed mean of M-value (blue: epithelial, red: endothelial). C, ACE2 expression is not regulated by inflammatory cytokines in HUVECs. qPCR analysis of ACE2 mRNA expression in HUVECs treated with a mix of 4 cytokines/chemokines (TNF-α, IL1-β, IL8, and IL6/IL6R chimeric protein) for 4 hours or 24 hours at 0, 0.01, 0.1, or 1.0 ng/mL. Data are normalized to GAPDH and presented as mean±SEM of 3 independent experiments. D and E, Very low-level, rare, and likely contaminating ACE2 transcripts are seen in ECs. D, ACE2 transcript reads are detected preferentially in PCs. UMAP landscapes of publicly available human heart data sets2 include 100 579 ECs, 77 856 PCs, 16 242 SMCs, and 718 MCs (https://www.heartcellatlas.org/). ACE2 transcript reads are detected preferentially in the PC cluster (enriching for ABCC9) and are rare in the EC cluster (enriching for PECAM1). E, PC transcripts are enriched together with ACE2 in 0.47% of ECs. Dot plot displaying the abundance of top-50 transcripts enriched ACE2+ versus ACE2- ECs, across cell types indicated in D. (The Wilcoxon rank-sum tests with Bonferroni-corrected P values are < 1E-60 for each). F, Epigenetic profiling indicates that the ACE2 gene is inactive in ECs. ChIP-seq binding profiles in HUVECs for histone modifications, RNA Pol2 enrichment, and deoxyribonuclease I hypersensitivity. The x axis represents the genomic position, the transcription start sites are indicated by closed arrow, and the direction of transcription is indicated by open arrows; the y axis shows ChIP-seq signal in reads per million per base pair (rpm/bp). The bottom row represents the chromatin state segmentation. Color key: active promoter, red; enhancers, yellow; transcriptional elongation, green; repressed, grey. G and H, Coronavirus replication in primary human cardiac and pulmonary ECs shows limited replication of SARS-CoV-2. G, Viral replication curves in human pulmonary (HPAEC) and cardiac (HCAEC) endothelial cells following infection with control HCoV-229E GFP (green fluorescent protein) reporter virus (MOI=0.6). Virus replication was measured via GFP fluorescence every 2 hours from 20 to 58 hours postinoculation. Mean±SEM of 3 technical replicates are shown at each time point for each biological replicate. H, Viral growth curves in HPAEC (n=3), HCAEC (n=3), and nonendothelial Vero cells (n=1) after infection with SARS-CoV-2 at MOI=10 or 100. Supernatant were collected at 1, 24, and 48 hours postinfection and virus copy number quantified by RT-qPCR detection of the SARS-CoV-2 N3 gene. ChIP-seq indicates chromatin immunoprecipitation sequencing; CPM, counts per million; EC, endothelial cell; HUVEC, human umbilical vein endothelial cell; IL, interleukin; MC, mesothelial cell; MOI, multiplicity of infection; PC, pericyte; qPCR, quantitative polymerase chain reaction; RT, real time; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SMC, smooth muscle cell; TCID, tissue culture infectivity dose; TNF, tumor necrosis factor; and UMAP, Uniform Manifold Approximation and Projection.
