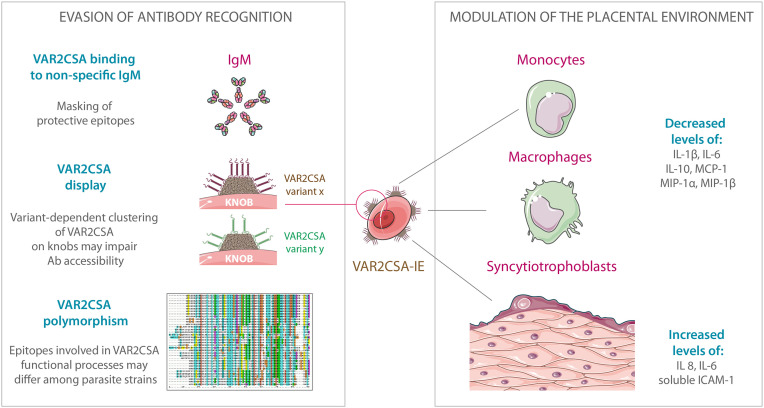
Figure 2.
Evasion of antibody recognition and modulation of the placental environment by VAR2CSA-expressing infected erythrocytes. IgM binding to VAR2CSA could mask protein epitopes recognized by anti-VAR2CSA IgGs and consequently alter opsonic phagocytosis of IEs. PfEMP1 clustering on knob structures may act as an immune evasion mechanism, impairing antibody accessibility to key residues involved in CSA-binding. Due to extensive polymorphism, epitopes involved in each VAR2CSA functional process may differ among parasite strains. Furthermore, multiplicity of var2csa genes within the parasite genome may also confer a greater capacity for antigenic variation and evasion of variant-specific immune responses. The presence of VAR2CSA on the IEs surface could lead to decreased production of IL-1β, IL-6, IL-10, MCP-1, MIP-1α, and MIP-1β by monocytes and macrophages. VAR2CSA-dependent binding of IEs to syncytiotrophoblasts is able to activate MAPK pathways and lead to increased secretion of IL-8, IL-6, and soluble ICAM-1. The art pieces used in this figure were modified from Servier Medical Art by Servier, licensed under a Creative Commons Attribution 3.0 Unported License (https://smart.servier.com/). The illustration of the protein sequence alignment is licensed under a Creative Commons Attribution, CC BY-SA 3.0 (https://creativecommons.org/licenses/by-sa/3.0/).