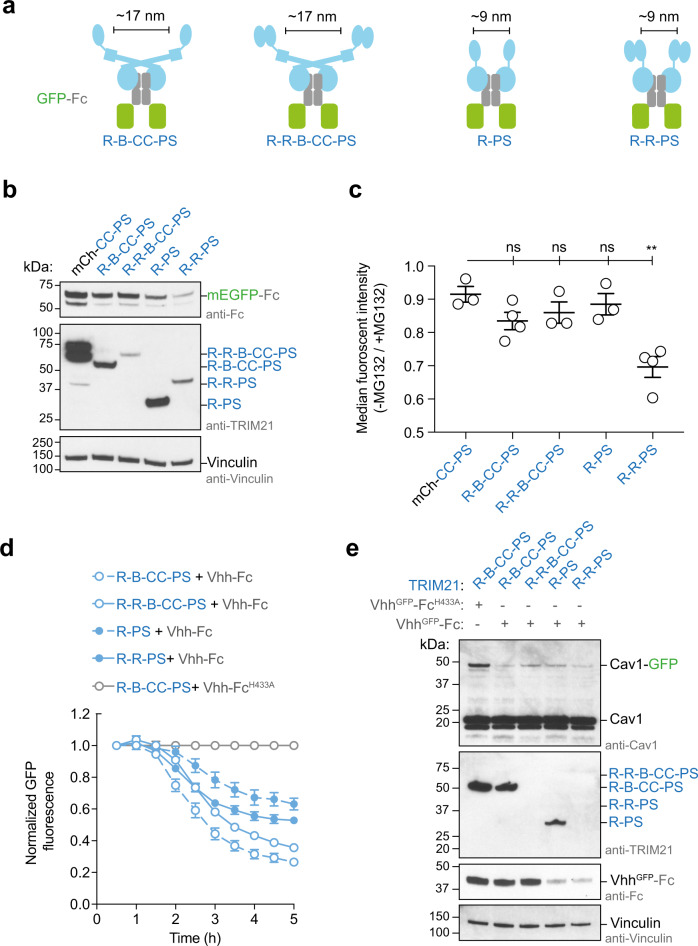
Fig. 5. Catalytic RING topology drives targeted protein degradation.
a Schematic cartoons showing the topology of TRIM21 (blue) on GFP-Fc (green and gray, respectively). b, c GFP-Fc degradation assay. b Western blot of RPE-1 TRIM21-knockout cells transiently expression GFP-Fc and a series of TRIM21 constructs. Western blots are representative of n = 2 independently performed experiments. c Shown is the flow cytometry analysis of green fluorescence of RPE-1 TRIM21-knockout cells transiently expressing GFP-Fc and a series of TRIM21 constructs. After electroporation, each population of cells was split in two and either treated with MG132 or DMSO. Data are presented as mean ± standard error of the mean. Each data point in the graph represents one biologically independently performed experiment (n = 3 (for mCh-CC-PS, R-R-B-CC-PS, and R-PS) or 4 (R-B-C-C-PS and R-R-PS)). A two-tailed unpaired Student’s T test was performed to assess the significance of fluorescence reduction relative to mCh-CC-PRYSPRY (P values: R-B-CC-PRYSPRY, 0.0797 (ns); R-R-B-CC-PRYSPRY, 0.02366 (ns); R-PRYSPRY, 0.4964 (ns); and R-R-PRYSPRY, 0.0035 (**)). d, e Trim-Away of Caveolin-1-mEGFP (Cav1-GFP) in NIH 3T3 GFP-Cav-1-knock in cells66. Shown in d is the normalized GFP fluorescence (error bars represent ± SEM of four images) and in e the western blot after the experiment. Data in d, e are representative of n = 2 independent experiments. Uncropped blots and raw data are provided in Source data. R RING, B Box, CC coiled-coil, PS PRYSPRY, mCh mCherry, kDa kilo Dalton, ns not significant.