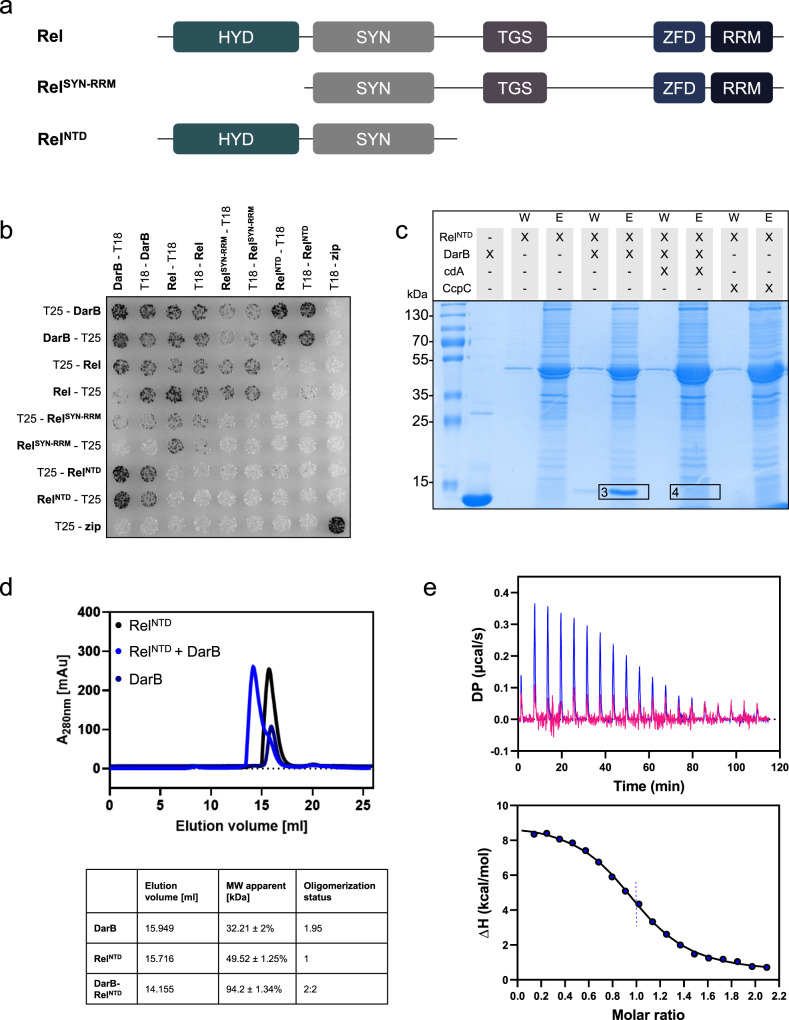
Fig. 2. DarB binds the N-terminal domain (NTD) of Rel.
a The domain organization of Rel and the truncated Rel variants used in this study. HYD, hydrolase domain; SYN, synthetase domain; TGS, TGS domain (for: ThrRS, GTPase, and SpoT); ZFD, a zinc finger domain; RRM domain (for RNA Recognition Motif). b Bacterial two-hybrid (BACTH) assay to test for the interaction between DarB and the full-length and truncated Rel-variants. N- and C-terminal fusions of DarB and the Rel variants to the T18 or T25 domain of the adenylate cyclase (CyaA) were created and the proteins were tested for interaction in E. coli BTH101. Dark colonies indicate an interaction that results in adenylate cyclase activity and subsequent expression of the reporter β-galactosidase. The experiment was conducted three times and a representative plate is shown. c In vitro pulldown experiment with the NTD of Rel. Strep-RelNTD was immobilized onto a StrepTactin column and incubated with DarB, DarB preincubated with c-di-AMP, or the control protein CcpC. The eluate and wash fractions were analyzed by SDS-PAGE and the presence of DarB in the elution fractions was further verified by MS analysis (excised gel bands are numbered with 3 and 4). The experiment was conducted three times and a representative gel is shown. d The DarB–RelNTD complex was analyzed by size exclusion chromatography and multi-angle light scattering (SEC-MALS). RelNTD and DarB were used in equimolar concentrations. Dark blue line, DarB; black line, RelNTD; blue line, mixture of DarB and Rel. The calculated molar masses determined by MALS are listed below the chromatogram. e The molar ratio of the DarB–RelNTD-complex was assessed by Isothermal titration calorimetry (ITC). The cell and the syringe contained 10 µM RelNTD and 100 µM DarB (blue) or 100 µM c-di-AMP-bound DarB (DarBcdA) (pink), respectively. cdA, c-di-AMP. Source data are provided as a Source data file.