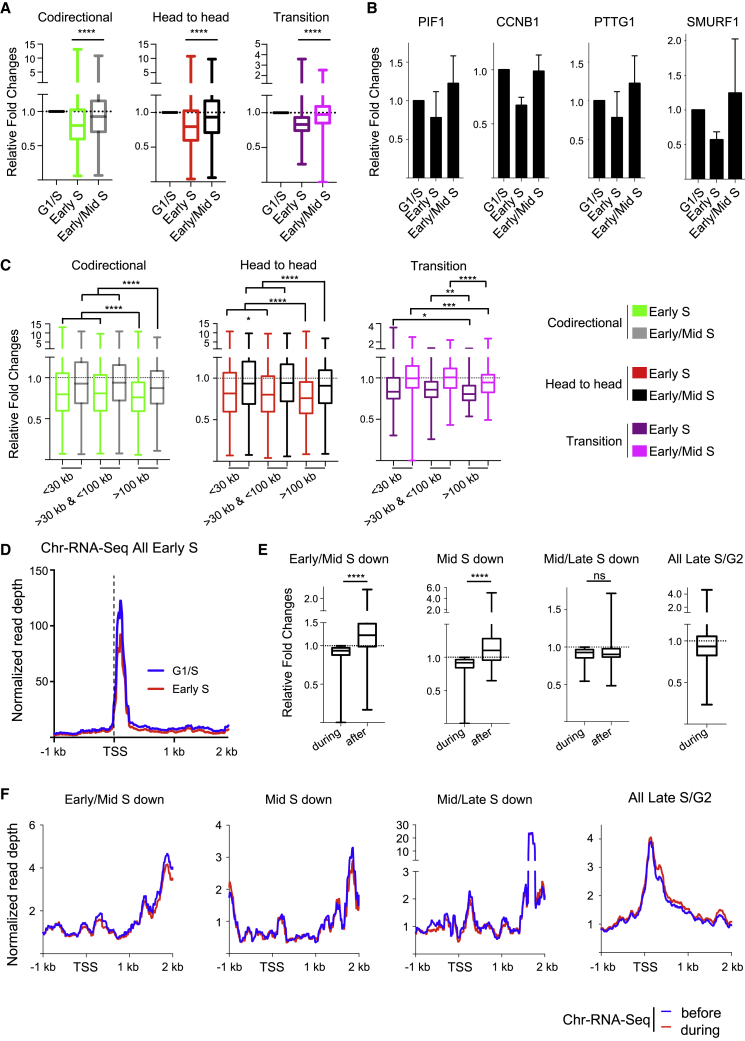
Figure 2.
Transient transcription shutdown when genes are replicated
(A) Quantification of Chr-RNA-seq levels over introns at the indicated time points, as fold change to G1/S. Genes are separated based on directionality as in Figure S1J.
(B) Nascent transcription levels in early S and early/mid-S compared to G1/S as in Figure 1D for genes replicated in early S; n = 3, data represent mean ± SEM.
(C) As in (A), with genes clustered by length.
(D) Average metagene profile of Chr-RNA-seq in G1/S and early S around the TSS −1 kb/+2 kb of genes replicated in early S.
(E) Fold change in Chr-RNA-seq levels for genes with reduced nascent transcription when replicated compared to the time point before, with levels during and after replication. The analysis includes 514 genes in early/mid-S, 144 genes in mid-S, 178 genes in mid-/late S, and all 716 genes in late S/G2; paired t test analysis.
(F) As in (D) but specifically over the genes highlighted in (E).
Box whiskers plots with line at the median, Mann-Whitney t test; ns, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.