Figure 4.
Persistence of RNAPII at TSSs prevents timely replication in S phase
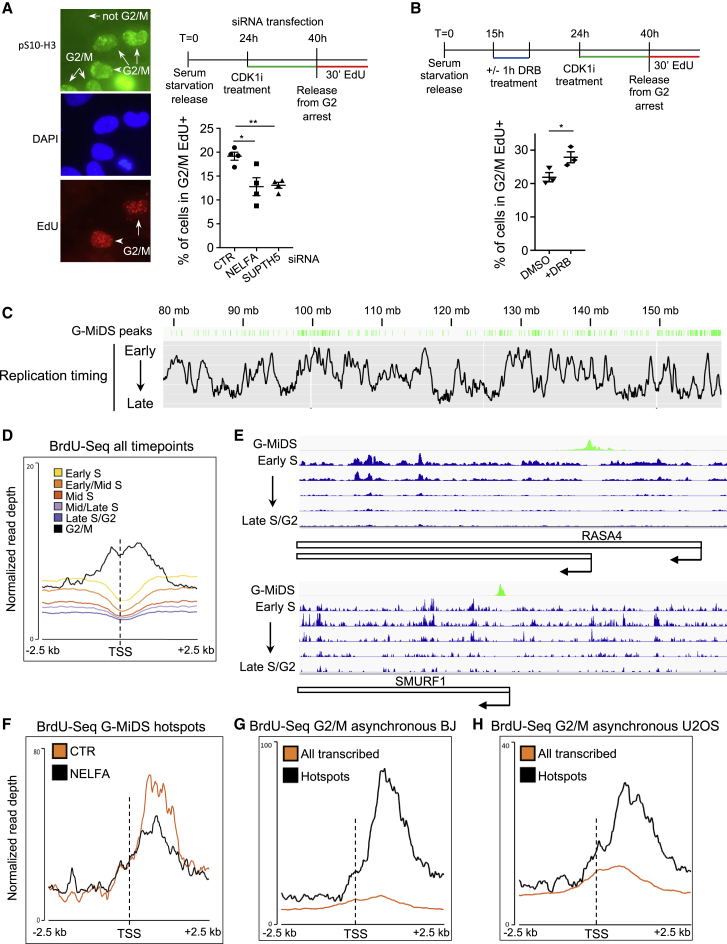
(A) Immunofluorescence for pS10-H3 (green) for G2/M-mitotic cells, EdU Click-iT for DNA synthesis (red), and DAPI (blue) for nuclei staining. G2/M-EdU, but not mitotic-EdU, double-positive cells quantified in CTR, NELFA, and SUPT5H siRNA cells; n = 4.
(B) G2/M-EdU double-positive cells quantified in CTR DMSO and cells treated in early S for 1 h with DRB (100 μM); n = 3.
(C) Distribution of G-MiDS-specific peaks in green in relation to replication timing on the long arm of chromosome 7, as in Figure S1A.
(D) Average metagene profile for BrdU-seq levels normalized to input DNA in S-phase time points and G2/M at TSSs ±2.5 kb of transcribed genes.
(E) Snapshots from IGV TDF (Integrative Genomics Viewer tiled data file) of G-MiDS-specific BrdU-seq (green) and BrdU-seq in S-phase time points (blue) around TSSs of the indicated genes.
(F) Average metagene profile for the BrdU-seq levels normalized to input DNA at TSS ±2.5 kb of the 449 hotspot genes in cells transfected with the denoted siRNA.
(G) As for (D) across all transcribed genes and only G-MiDS hotspots for asynchronous BJ cells treated with BrdU for 30 min and sorted in G2/M.
(H) As for (G), with U2OS cells.
Data represent mean ± SEM; Student’s t test; ∗p < 0.05; ∗∗p < 0.01.